Cells communicate with each other via receptor-ligand interactions. Researchers at Stanford University have developed lentiviral-mediated cell entry by engineered receptor-ligand interaction (ENTER) to display ligand proteins, deliver payloads, and record receptor specificity. They optimize ENTER to decode interactions between T cell receptor (TCR)-MHC peptides, antibody-antigen, and other receptor-ligand pairs. A viral presentation strategy allows ENTER to capture interactions between B cell receptor and any antigen. The researchers engineer ENTER to deliver genetic payloads to antigen-specific T or B cells to selectively modulate cellular behavior in mixed populations. Single-cell readout of ENTER by RNA sequencing (ENTER-seq) enables multiplexed enumeration of antigen specificities, TCR clonality, cell type, and states of individual T cells. ENTER-seq of CMV-seropositive patient blood samples reveals the viral epitopes that drive effector memory T cell differentiation and inter-clonal vs. intra-clonal phenotypic diversity targeting the same epitope. ENTER technology enables systematic discovery of receptor specificity, linkage to cell fates, and antigen-specific cargo delivery.
ENTER-seq for massively parallel measurement of
antigen peptide sequence, TCR sequence, and transcriptome
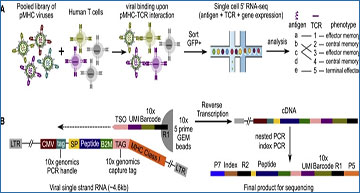
(A) Schematic view of ENTER-seq workflow. A library of pooled viruses displaying individual pMHC was incubated with T cells for 2 h. GFP+ T cells were sorted for droplet-based single-cell genomics profiling. (B) Viral RNA engineering strategy for droplet-based single-cell capture. 10X Genomics capture tag is inserted in the linker region between B2M and HLA-A2. 10X Genomics PCR handle is inserted after CMV promoter. CMV, CMV promoter; SP, signal peptide sequence; peptide, antigen peptide; B2M, beta-2-microglobulin; MHC class I, HLA-A0201 allele; LTR, long terminal repeat; TSO, template-switching oligo sequence. (C) Schematic view of T cell mixing experiment for ENTER-seq. (D) Number of pp65495–503-TCR T cells UMI counts (x axis) and NY-ESO-1157–165-TCR UMI counts (y axis) associated with each cell barcode (dot). The colors are assigned as NY-ESO-1157–165-TCR+ T cells (light blue), pp65495–503-TCR+ T cells (red), and doublets (green, with both NY-ESO-1157–165-TCR and pp65495–503-TCR). (E) Scatterplots of TCR UMI counts after doublet removal, colored by enrichment ratio of pp65495–503-antigen UMI count among total UMI counts (left) and enrichment ratio of NY-ESO-1157–165-antigen UMI count among total UMI counts (right). (F) Number of pp65(495–503)-antigen UMI counts (x axis) and NY-ESO-1157–165-antigen UMI counts (y axis) associated with each cell barcode (dot) after doublet removal. The colors are assigned as NY-ESO-1157–165-TCR+ T cells (light blue) and pp65495–503-TCR+ T cells (red).
Yu B, Shi Q, Belk JA, Yost KE, Parker KR, Li R, Liu BB, Huang H, Lingwood D, Greenleaf WJ, Davis MM, Satpathy AT, Chang HY. (2022) Engineered cell entry links receptor biology with single-cell genomics. Cell [Epub ahead of print]. [article]