Tumor development is a multistep process that involves many genetic alterations that lead to cell growth and proliferation. As the tumor evolves, substantial crosstalk with various cell types, including non-cancer cells from the surrounding microenvironment, facilitates tumor growth and development. Traditionally, guidelines for treating cancer have been based on staging the tumor using anatomical features such as tumor size or histological grade. However, these classifications do not consider the surrounding tumor microenvironment; therefore, they are insufficient to reflect prognosis and predict treatment response. With the development of transcriptomic technologies that profile gene expression in situ with spatial context, it is now possible to evaluate the utility of RNA or protein expression signatures for tumor samples with varying heterogeneity.
Over the past decade, transcriptomic technologies such as bulk tissue RNA-Seq have been widely used to understand variations in gene expression across different tissues and disease conditions. This has enabled the discovery of numerous gene expression signatures (GESs), defined as a list of genes whose differential expression is associated with a specific biological process such as tumor progression. Numerous GESs have been applied to various cancers to classify tumor types, identify tumor stages, and predict disease prognosis. Prognostic GESs have helped guide treatment decisions for certain types of cancers, such as breast cancer, wherein microarray-based assays are used to assign tumors into categories of low or high risk of metastasis or of different types of tumors such as Luminal-, HER2- or Basal-type. However, no signature has been adopted in routine clinical practice in colorectal cancer despite many gene expression profiling studies on prognosis having been performed.
A remarkable feature of colorectal cancer is the difference in prognosis between the early and late stages. Early stages I and II exhibit a moderate risk of relapse after surgical resection, whereas stage III has a higher chance of recurrence. Moreover, colorectal cancers are often diagnosed and detected at an advanced stage. This has propelled the discovery of several GESs for colorectal cancer that show significant statistical association with prognosis. However, their ability to accurately classify independent samples into high-risk and low-risk groups has been limited. A recent publication by Fisher et al. in Clinical Cancer Research suggests that many GESs can be misinterpreted when applied to complex heterogenous tumor biopsies, making them ineffective in distinguishing colorectal tumor phenotypes (1). One of the primary reasons for this discrepancy is that precise mechanism-based GESs, such as those with an epithelial cell focus to detect epithelial–mesenchymal transitions (EMT), may no longer reflect epithelial-specific gene expression when applied to a heterogeneous tumor sample. EMT is a developmental program that enables tumor cells to suppress their epithelial features and transform into mesenchymal cells, thus gaining the ability to migrate from the primary site of the tumor. Such specific GESs often reflect phenotypic changes under tightly controlled experimental conditions of an in vitro model system. However, applying these specific GESs to bulk RNA-Seq data from heterogenous tumors fails to stratify tumors based on the phenotype it was designed for. In the aforementioned study, the misinterpretation of GESs was shown to be due to the relatively high expression levels of the GES in nonepithelial cells of the stroma, thus preventing the detection of the phenotypic change in epithelial cells.
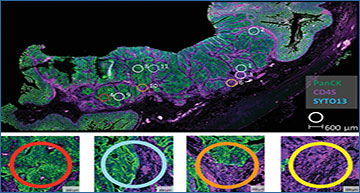
Using colorectal cancer as an example, the study by Fisher et al. used orthogonal profiling methods, including spatial transcriptomics, to comprehensively assess the contribution of stroma to some of the most widely used GESs in cancer research. About 8,000 commonly employed GESs were assessed from a bulk tumor dataset obtained from 360 patients (the FOCUS Cohort). The data was further independently validated using laser capture microdissection, flow cytometry, and single-cell RNA-Seq methods that distinguish epithelium and stroma from the tumor mass. For in situ spatial transcriptomic profiling, the unique ability of NanoString’s GeoMx® Digital Spatial Profiler (DSP) was used to look at gene expression from a colorectal cancer biopsy from different tissue regions defined by fluorescent staining of morphological markers. Immunofluorescence expression of pan-cytokeratin (PanCK) distinguished the epithelium and stromal compartments and helped guide the selection of numerous regions of interest (ROIs) corresponding to regions representing the epithelial tumor center, tumor microenvironment, and interface between the tumor and tumor microenvironment. Each of the selected ROIs was profiled separately. Upon analysis, as expected, there was enrichment in the EMT signaling cascade, specifically in the stromal (PanCK-) regions, which was orders of magnitude higher than that of the epithelial cells. Consequently, if typical GESs for colorectal cancer were applied to bulk transcriptomic data sets, the relative amount of expression changes in the stroma would overwhelm and overshadow the rather subtle epithelial transitions in the tumor, thus skewing the interpretation of the GES. Using the FOCUS cohort as a comparison, there was excellent concordance in single set Gene Set Enrichment Analysis (ssGSEA) scores of the complete molecular signature database (MSigDB) hallmark EMT signature (r = 0.95; n = 200 genes) and MYC Targets V2 signature (r = 0.75; n = 58 genes), when assessed using corresponding signatures present in the GeoMx® Cancer Transcriptome Atlas.

Eleven geometric regions of interest (ROIs) corresponding to epithelial tumor center, abundant tumor microenvironment (TME) regions, and regions representing an interface between tumor and TME were selected for spatial transcriptomics with the GeoMx Cancer Transcriptome Atlas based on positive or negative staining for PanCK. Figure reproduced with permission from Fisher NC al. Clinical Cancer Research. 2022 under the Creative Commons license.
The use of GeoMx DSP in the Fisher et al. study demonstrates the importance of profiling methods that incorporate tissue heterogeneity in addition to transcriptional signatures. Although bulk tumor datasets are high-throughput and can quantify gene expression levels across a large population of cells, the nature of bulk profiling obscures inherent biological differences between different tissue compartments and cell types. Single cell-sequencing (scRNA-Seq) methods attempt to address this heterogeneity by providing high-quality transcriptional profiling of individual cells; however, analyzing isolated cells away from their native environment ignores key spatial context that is crucial to biological function and crosstalk. As the authors themselves noted: “while bulk tumor datasets will remain an essential tool for statistical association studies, these data clearly highlight the need for the compartment and/or lineage-specific stratification, as afforded by ST (spatial transcriptomics), to ensure accurate biological interpretation of GESs.” Thus, spatial profiling with the GeoMx DSP becomes an invaluable tool to augment biological findings from traditional bulk RNA-Seq studies.
References
- Fisher NC, Byrne RM, Leslie H, Wood C, Legrini A, Cameron AJ, Ahmaderaghi B, Corry SM, Malla SB, Amirkhah R, McCooey AJ, Rogan E, Redmond KL, Sakhnevych S, Domingo E, Jackson J, Loughrey MB, Leedham S, Maughan T, Lawler M, Sansom OJ, Lamrock F, Koelzer VH, Jamieson NB, Dunne PD. Biological Misinterpretation of Transcriptional Signatures in Tumor Samples Can Unknowingly Undermine Mechanistic Understanding and Faithful Alignment with Preclinical Data. Clin Cancer Res. 2022 Sep 15;28(18):4056-4069. doi: 10.1158/1078-0432.CCR-22-1102. PMID: 35792866; PMCID: PMC9475248.
- Sanz-Pamplona R, Berenguer A, Cordero D, Riccadonna S, Solé X, Crous-Bou M, Guinó E, Sanjuan X, Biondo S, Soriano A, Jurman G, Capella G, Furlanello C, Moreno V. Clinical value of prognosis gene expression signatures in colorectal cancer: a systematic review. PLoS One. 2012;7(11):e48877. doi: 10.1371/journal.pone.0048877. Epub 2012 Nov 7. PMID: 23145004; PMCID: PMC3492249.