Understanding how cells work at a fundamental level is crucial for advancing medical and biological sciences. One of the lesser-explored areas of cellular biology is the mechanical properties of individual cells and how these properties are regulated at the molecular level. A new technique called ELASTomics developed by researchers at the RIKEN Institute promises to open up this field by allowing scientists to study the mechanical state of cells alongside their genetic activity. Let’s break down this exciting development.
What is ELASTomics?
ELASTomics stands for Electroporation-based Lipid-Bilayer Assay for cell Surface Tension and transcriptomics. This method allows researchers to study both the mechanical properties of a cell’s surface and its genetic activity at the same time. By doing so, scientists can gain a deeper understanding of how cells function and respond to different conditions.
Workflow of ELASTomics
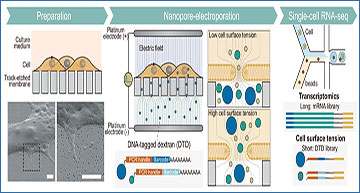
(Preparation) Cells are cultured on the track-etched membrane coated by fibronectin. The lower left panel is field emission scanning electron microscopy (FE-SEM) images of MCF7 cells on a track-etched membrane, and the lower right panel is zoom-up views of the dotted box in the lower left panel. An independent duplicate experiment has verified similar results at a different angle of FE-SEM imaging. Scale bars, 2 μm. (Nanopore-electroporation) Cells are subjected to pulsed electric fields that are created in the vicinity of nanopores to import DNA-tagged dextran (DTD) into the cells. The cell surface tension of the lipid bilayer affects the diameter of the pore formed in a lipid bilayer. The amount of imported DTD thus dictates the surface tension of individual cells. (Single-cell RNA-seq) Single-cell RNA-sequencing (scRNA-seq) workflow creates libraries of transcriptomics (mRNA library) and cell surface tension (DTD library)
How Does ELASTomics Work?
The technique involves using nanopore electroporation, a process that temporarily opens tiny pores in the cell membrane. Through these pores, specially labeled molecules can be introduced into the cell. These molecules can help measure the tension on the cell’s surface, which is an indicator of its mechanical state. Additionally, the same molecules can be used to analyze the cell’s RNA, giving insights into the cell’s genetic activity.
Integration with Existing Technologies
One of the key advantages of ELASTomics is its compatibility with existing single-cell sequencing technologies. This means that researchers can easily combine this new method with tools they are already using, making it easier to adopt and integrate into current research workflows.
Validating ELASTomics
To ensure that ELASTomics works as intended, the researchers tested it on various cancer cell lines. The results were promising, showing that the method could accurately identify different cell types based on their surface tension and genetic activity. This validation is crucial because it demonstrates that ELASTomics can provide reliable data for further research.
Exploring Cellular Mechanics
ELASTomics has been used to study the relationship between cell surface tension and the genetic regulation of cells. For instance, researchers looked at how cell surface mechanics change during cellular aging, a process known as senescence. They discovered that a protein called RRAD plays a role in regulating the surface tension of senescent cells, providing new insights into the aging process.
Applications and Future Potential
The ability to profile both mechanical and molecular characteristics of cells opens up many exciting possibilities. For example, understanding how cell mechanics influence cancer progression could lead to new treatments. Similarly, insights gained from studying cellular aging could inform strategies to combat age-related diseases.
Shiomi A, Kaneko T, Nishikawa K, Tsuchida A, Isoshima T, Sato M, Toyooka K, Doi K, Nishikii H, Shintaku H. (2024) High-throughput mechanical phenotyping and transcriptomics of single cells. Nat Commun 15(1):3812. [article]