Image by kan_khampanya/Shutterstock
A research group led by University of Tsukuba has discovered a novel RNA viral genome from microbes inhabiting a high-temperature acidic hot spring. Their study shows that RNA viruses can live in high-temperature environments (70℃-80℃), where no RNA viruses have been observed before. In addition to the two known RNA virus kingdoms, a third kingdom may exist.
There are numerous RNA virus species on Earth. However, their diversity and evolution as well as roles in the ecosystem remain unclear.
In this study, using an original method, researchers have discovered a novel RNA viral genome from thermoacidophilic microbes (close to the last universal common ancestor of life) in the hot springs of Unzen and Kirishima fumaroles. This RNA virus was named hot spring RNA virus (HsRV) and was presumed to infect thermoacidophilic bacteria. This study shows that RNA viruses can inhabit high-temperature environments, where life is believed to have originated. Furthermore, HsRV considerably differs from all other RNA viruses belonging to the two established RNA virus kingdoms, indicating the existence of a previously overlooked third RNA virus kingdom.
A thermoacidophilic partiti-like virus
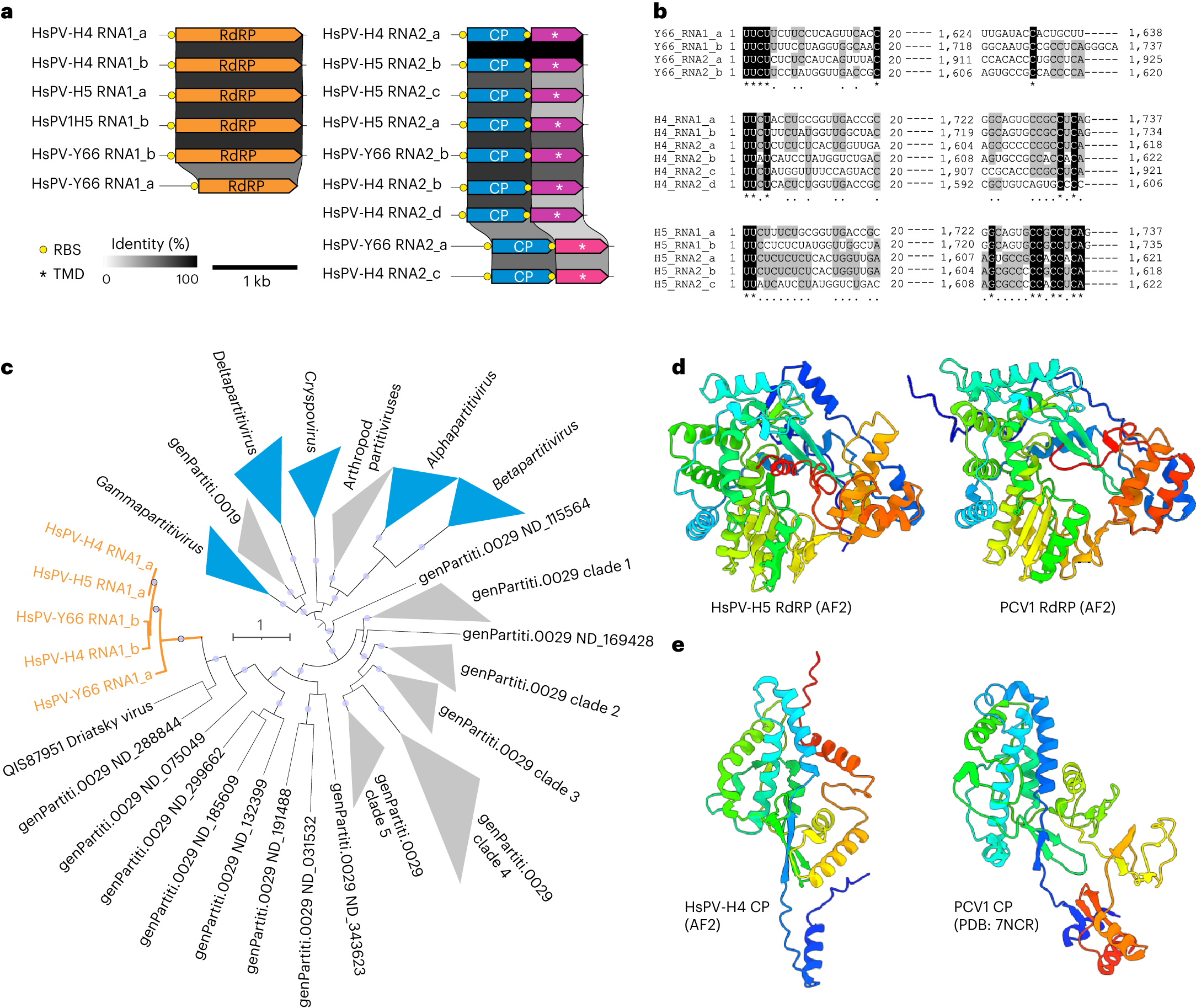
a, Genome organization and conservation of the two genome segments, RNA1 and RNA2, of HsPV. ORFs encoding homologous proteins are shown as arrows with identical colours. Yellow circles represent predicted SD RBS. Asterisks denote putative genes encoding predicted TMD-containing proteins. b, MSA of the 5ʹ- and 3ʹ-terminal regions of the coding strands of reconstructed genome segments. Black shading, 100% nucleotide identity; grey shading, >50% nucleotide identity. c, Maximum-likelihood phylogeny of the RdRP proteins from representative members of the family Partitiviridae and related sequences (including all HsPV strains, shown in orange). Clades corresponding to the official Partitiviridae genera are shown in blue, whereas those corresponding to unclassified groups are in grey. Node supports were assessed using the SH-aLRT; circles indicate nodes with ≥90% supports. The scale bar represents the number of substitutions per site. d, Comparison of the RdRP from HsPV-H5 with a homologue from deltapartitivirus PCV1. e, Comparison of the CP from HsPV-H4 with a homologue from deltapartitivirus PCV1. The structures are coloured using the rainbow scheme, from blue N terminus to red C terminus.
Future studies will attempt to culture host strains that harbor HsRV and elucidate the virological properties and ecology of HsRV. In addition, the same method used in this study will be applied to various microorganisms, animals, and plants to explore possible undiscovered RNA viruses.
Urayama SI, Fukudome A, Hirai M, Okumura T, Nishimura Y, Takaki Y, Kurosawa N, Koonin EV, Krupovic M, Nunoura T. (2024) Double-stranded RNA sequencing reveals distinct riboviruses associated with thermoacidophilic bacteria from hot springs in Japan. Nat Microbiol [Epub ahead of print]. [article]