Single-cell sequencing provides detailed insights into biological processes including cell differentiation and identity. While providing deep cell-specific information, the method suffers from technical constraints, most notably a limited number of expressed genes per cell, which leads to suboptimal clustering and cell type identification.
Researchers at the University Medical Center Hamburg-Eppendorf have developed DISCERN, a novel deep generative network that precisely reconstructs missing single-cell gene expression using a reference dataset. DISCERN outperforms competing algorithms in expression inference resulting in greatly improved cell clustering, cell type and activity detection, and insights into the cellular regulation of disease. The researchers show that DISCERN is robust against differences between batches and is able to keep biological differences between batches, which is a common problem for imputation and batch correction algorithms. They use DISCERN to detect two unseen COVID-19-associated T cell types, cytotoxic CD4+ and CD8+ Tc2 T helper cells, with a potential role in adverse disease outcome. They utilize T cell fraction information of patient blood to classify mild or severe COVID-19 with an AUROC of 80% that can serve as a biomarker of disease stage. DISCERN can be easily integrated into existing single-cell sequencing workflow.
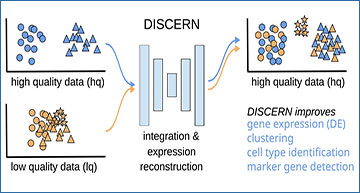
Integration and expression reconstruction of single-cell sequencing data
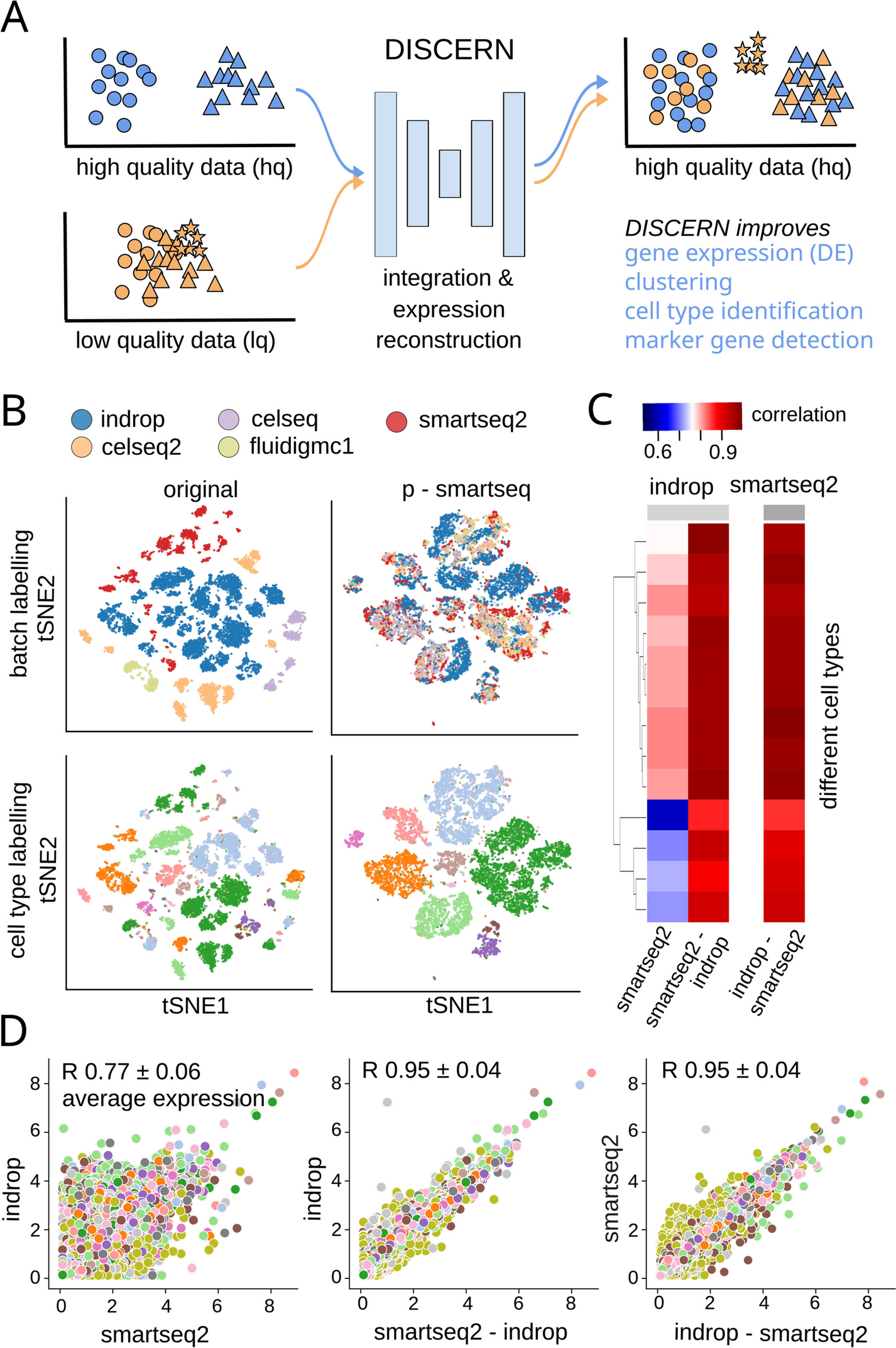
A DISCERN transfers the style of a high-quality (hq) dataset to a related low-quality (lq) dataset, enabling gene expression reconstruction that results in improved clustering, cell type identification, marker gene detection, and mechanistic insights into cell function. The hq and lq datasets have to be related but not identical, containing for example several overlapping cell types but also exclusive cell types of cell activity states for one or the other dataset. B t-SNE visualization of the pancreas dataset before reconstruction (original) and after transferring the style of the smartseq2 dataset using DISCERN (p-smartseq2). The upper row shows the dataset of origin before and after reconstruction colored by batch and the lower row colored by cell type annotation (details of 13 cell types in supplements). C and D Average gene expression (over all the cells of a given type) of the pancreas indrop and smartseq2 datasets before (first column and panel) and after smartseq2 to indrop (second column and panel) and after indrop to smartseq2 reconstruction (third column and panel). C Gene correlation by cell type shown in colored heatmap. D Each colored point represents a single gene colored by the cell type. The mean Pearson correlation with one standard deviation over all cell types is shown in the figure title
DISCERN is a flexible tool for reconstructing missing single-cell gene expression using a reference dataset and can easily be applied to a variety of data sets yielding novel insights, e.g., into disease mechanisms.
Availability – All original code has been deposited at https://github.com/imsb-uke/discern and https://zenodo.org/record/8260693 and is publicly available under the MIT License.
Hausmann F, Ergen C, Khatri R, Marouf M, Hänzelmann S, Gagliani N, Huber S, Machart P, Bonn S. (2023) DISCERN: deep single-cell expression reconstruction for improved cell clustering and cell subtype and state detection. Genome Biol 24(1):212. [article]