A cutting edge technique, called Direct RNA sequencing, shows that ‘jumping genes’ could be the key to help plants become more resilient.
‘Jumping genes, or transposons, are sections of DNA that can copy themselves and jump between different parts of the genome, and might help plants adapt to stressful, changing conditions, finds a study published in Nature Communications on June 5, 2023.
Using a cutting-edge sequencing technique, researchers from the Okinawa Institute of Science and Technology (OIST) and the Center for Sustainable Resource Science, RIKEN found that Arabidopsis thaliana, a plant used as a model for scientific research, expresses thousands of hybrids between regular genes and jumping genes. The plant alters the expression of these hybrid genes in response to environmental stresses such as excessive heat, or pathogens. These findings could contribute to the development of new crops that can better cope with stressful environments.
Regulation of gene expression (in other words, how a cell controls which genes are active) is vital for the correct functioning of all living things. When genes are expressed, genetic instructions are converted from DNA into RNA transcripts, ensuring useful products like proteins are made at the right time and in the right place.
Typically, plants and animals also use gene regulation to suppress the activity of transposons.
Dr. Jeremy Berthelier, lead author of the study and post-doctoral researcher in the Plant Epigenetics Unit, explained: “Transposons can be damaging, as these sections of DNA can insert themselves into genes and cause harmful mutations.”
However, in this new study, the scientists found that Arabidopsis thaliana expresses thousands of RNA transcripts that are hybrids of regular genes and transposons. These hybrid RNA molecules, called gene-transposon transcripts, occur when transposons jump near or within a gene, and are then expressed together.
The researchers identified these transcripts using a cutting-edge method called Direct RNA Sequencing that can read long RNA sequences. They then used a computational tool they developed, called ParasiTE, to classify the gene-transposon transcripts, based on the effect that the transposon had on the gene.
Detection of TE-gene transcripts (TE-G transcripts) and alternative TE-gene isoforms
(ATE-G isoforms) in DRS-AtRTD3 of the Arabidopsis transcriptome
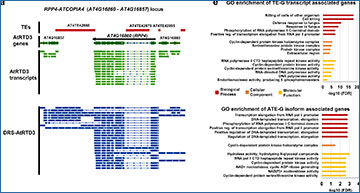
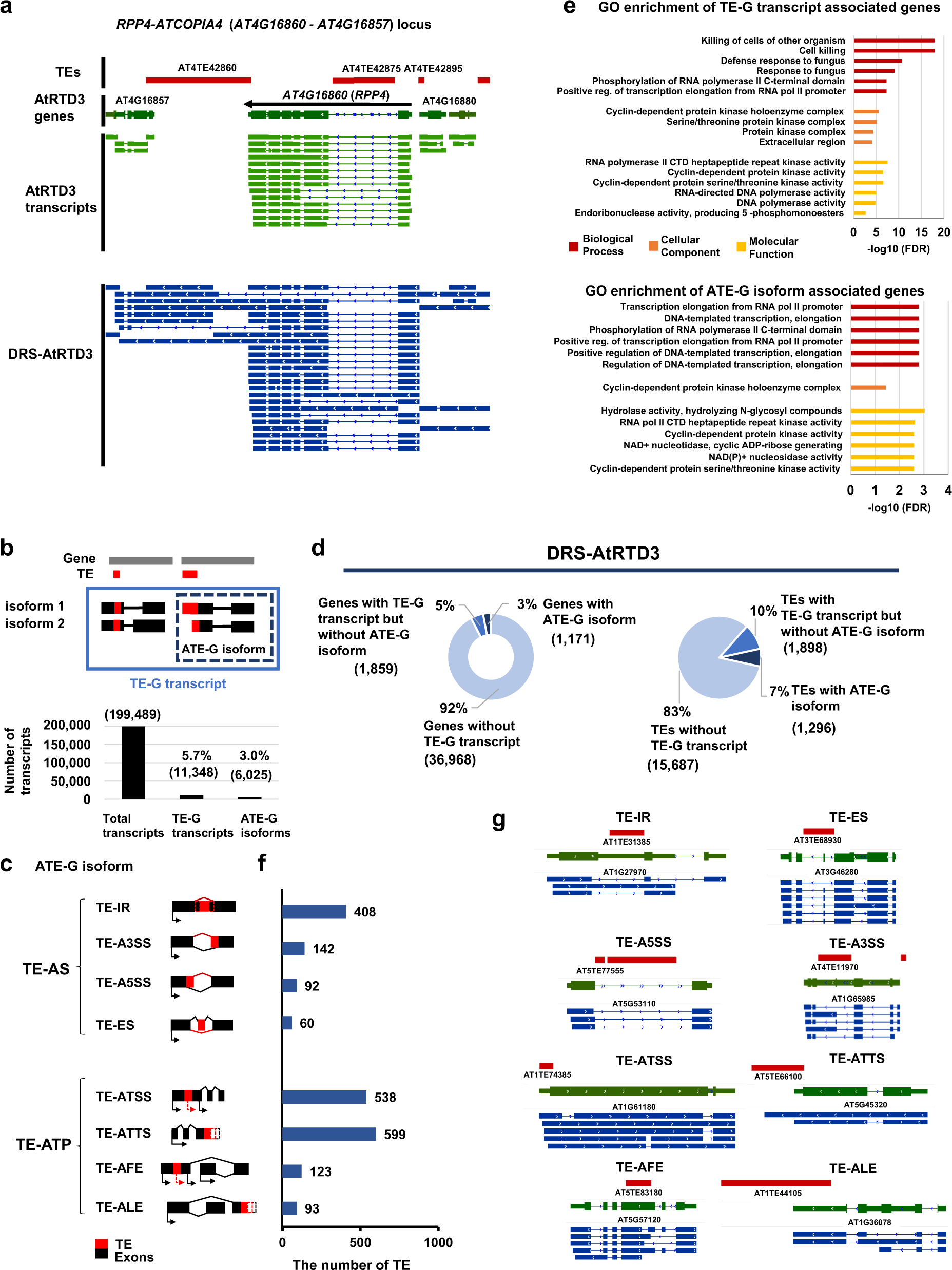
a Representative TE-G transcripts associated with the RPP4-ATCOPIA4 locus detected in DRS-AtRTD3. b The number of TE-G transcripts and ATE-G isoforms detected by ParasiTE. c RNA processing events associated with ATE-G isoforms (TE-AS and TE-ATP). TE-AFE and TE-ALE are included in TE-ATSS and TE-ATTS, respectively. AS; Alternative Splicing, ATP; Alternative Transcript Production, IR; Intron Retention, ES; Exon Skipping, A5SS; Alternative 5′ Splicing Sites, A3SS; Alternative 3′ Splicing Sites, ATSS; Alternative Transcription Start Sites, ATTS; Alternative Transcription Termination Sites, AFE; Alternative First Exon, ALE; Alternative Last Exon. d The number of genes (left; 39,998 gene models based on DRS-AtRTD3 annotation; 27,628 genes were associated with Arabidopsis Genome Initiative codes) and TEs (right; n = 18,881 TE annotations) associated with TE-G transcripts and ATE-G isoforms identified in DRS-AtRTD3. e Enriched Gene Ontology terms of genes associated with TE-G transcripts (top) and ATE-G isoforms in DRS-AtRTD3 (bottom). f The number of TEs associated with ATE-G isoform events. Some TEs were included in several ATE-G isoform categories. g Representative loci associated with TE-AS and TE-ATP events detected in DRS-AtRTD3. Red, TE annotation; Green, AtRTD3 annotation (collapsed); Blue, DRS-AtRTD3 annotation (extended). Source data are provided as a Source Data file.
The researchers then conducted a systematic study of how environmental stresses affect gene-transposon transcripts. They found that a transposon called ONSEN, caused a change in expression of its associated gene, GER5, in response to excessive heat.
Another finding relates to a gene called RPP4, which produces a protein that helps A. thaliana fight against pathogen infections. The researchers found that suppressing expression of the gene-transposon transcripts of the RPP4 gene affects the plant’s resistance to pathogens.
Gene-transposon transcripts can therefore help plant adapt to environmental stresses and changing environmental conditions.
“The broader implications of the study are that transposons can regulate their associated genes in a sophisticated way, by changing their DNA sequence and modulating their expression and stability. This may be very important for plant life, and their response to changing environmental conditions, such as extreme temperatures or to pathogens,” explained Dr. Berthelier.
The study could also prove to be useful for developing resilient crops.
Professor Hidetoshi Saze, senior author of the study and head of the Plant Epigenetic Unit at OIST, said “We can now study other plants that are important crop species. We could find even more genes which have transposon-dependent regulation and target these genes for editing , to make more productive crops for cultivation.”
Source – Okinawa Institute of Science and Technology
Berthelier J, Furci L, Asai S, Sadykova M, Shimazaki T, Shirasu K, Saze H. (2023) Long-read direct RNA sequencing reveals epigenetic regulation of chimeric gene-transposon transcripts in Arabidopsis thaliana. Nat Commun 14(1):3248. [article]