A major informatic challenge in single cell RNA-sequencing analysis is the precise annotation of datasets where cells exhibit complex multilayered identities or transitory states. Researchers at the Stanford University School of Medicine have developed devCellPy a highly accurate and precise machine learning-enabled tool that enables automated prediction of cell types across complex annotation hierarchies. To demonstrate the power of devCellPy, the researchers construct a murine cardiac developmental atlas from published datasets encompassing 104,199 cells from E6.5-E16.5 and train devCellPy to generate a cardiac prediction algorithm. Using this algorithm, they observe a high prediction accuracy (>90%) across multiple layers of annotation and across de novo murine developmental data. Furthermore, they conduct a cross-species prediction of cardiomyocyte subtypes from in vitro-derived human induced pluripotent stem cells and unexpectedly uncover a predominance of left ventricular (LV) identity that they confirmed by an LV-specific TBX5 lineage tracing system. Together, these results show devCellPy to be a useful tool for automated cell prediction across complex cellular hierarchies, species, and experimental systems.
Overview of devCellPy
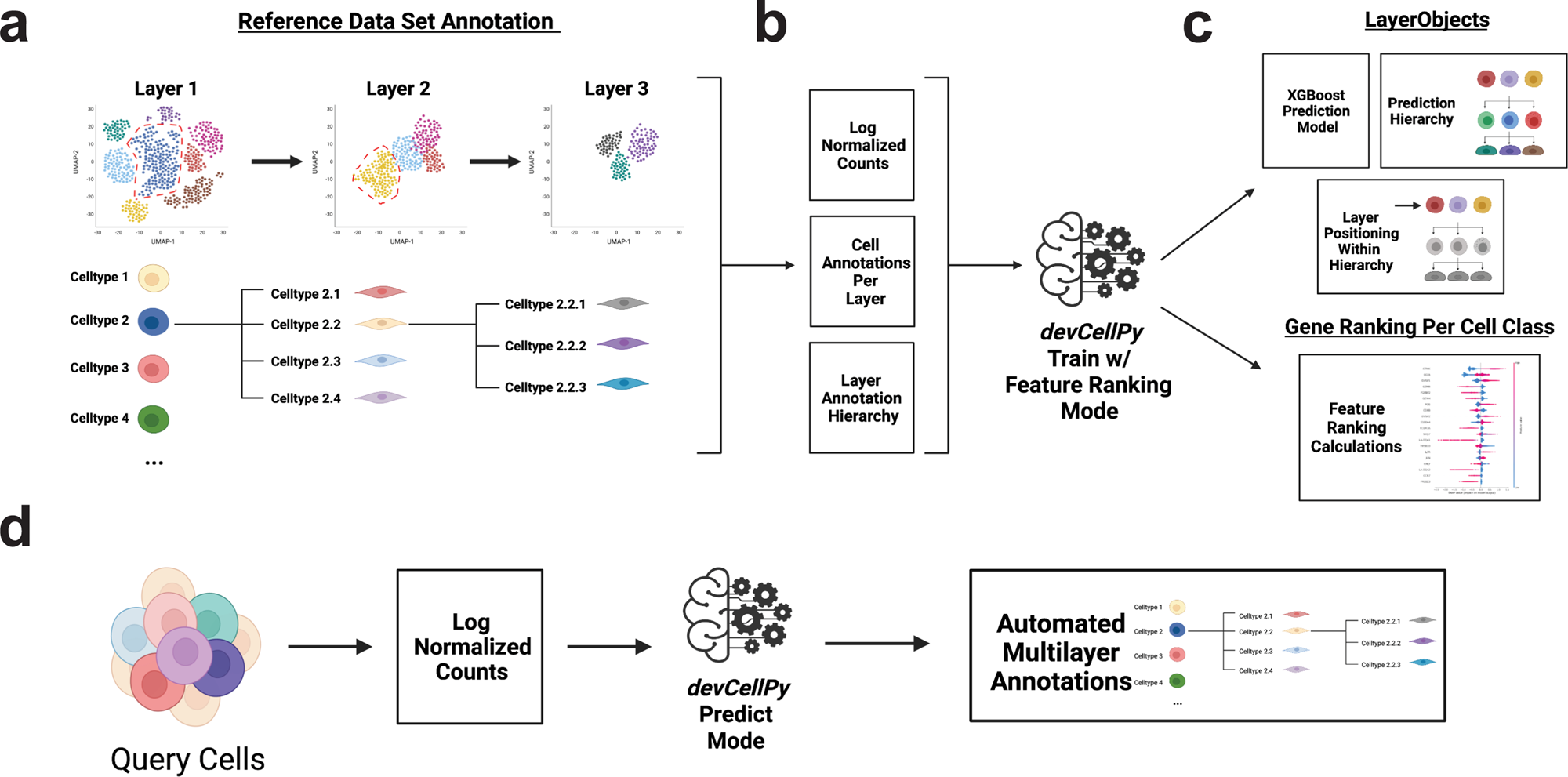
devCellPy is a multilayered machine learning algorithm for the hierarchical annotation of single-cell RNA-seq data. a Reference data is constructed by conducting annotation of the dataset across multiple layers. These layers follow a hierarchical structure where cell subtype annotations are nested within broader categories. The hierarchical structure can be present for any combination of layers and sublayers. b To train devCellPy, log normalized counts, individual cell annotations per layer, and a layer annotation hierarchy are fed into the algorithm’s train mode. c devCellPy will create a LayerObject for each layer of annotation within the hierarchy. The LayerObject consists of an XGBoost prediction model that is trained on the reference data and will also encode the position of the layer within the annotation hierarchy. LayerObjects allow for the automated prediction of cell types across all layers of annotation. Importantly, devCellPy follows the hierarchy’s organizational logic meaning that cell subtypes will be predicted only if they fall within a specified branch within the hierarchy. Under the feature ranking mode, devCellPy will use the SHAP algorithm to determine the positive and negative gene predictors per cell type annotated. d devCellPy can conduct automated prediction of query cells by exporting log normalized counts and feeding the matrix to the algorithm. The output will contain an automated annotation across multiple layers.
Availability -The code used for the generation of devCellPy is available at https://github.com/devCellPy-Team/devCellPy.
Galdos FX, Xu S, Goodyer WR, Duan L, Huang YV, Lee S, Zhu H, Lee C, Wei N, Lee D, Wu SM. (2022) devCellPy is a machine learning-enabled pipeline for automated annotation of complex multilayered single-cell transcriptomic data. Nat Commun 13(1):5271. [article]