By Patrick Danaher, PhD – Principal Biostatistician, NanoString Technologies
While admitting to some bias, I’ll assert that CosMx Spatial Molecular Imager (SMI) data is flat-out awe-inspiring: even a perfunctory analysis of a single run produces a terabyte of data, gorgeous images (see below), and spatial relationships from the scale of centimeters to micrometers that you could spend weeks exploring.
Spatial Molecular Imager (SMI) data is flat-out awe-inspiring: even a perfunctory analysis of a single run produces a terabyte of data, gorgeous images (see below), and spatial relationships from the scale of centimeters to micrometers that you could spend weeks exploring.

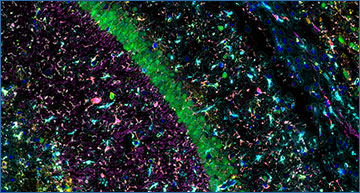
This is a tissue section from a Mouse Hippocampus that has been run on the CosMx SMI with the new CosMx Mouse Neuroscience Protein Panel. Multianalyte markers employed allow for superior on-platform cell segmentation, providing the most accurate single-cell and subcellular transcript information.
Marker Key: DAPI Iba1NeuN ALDH1L1 GFAP GAD67 P2RY12 ChAT
But ultimately, you don’t need amazing images, you need answers to biological questions. Here I’ll lay out questions typically asked with spatial transcriptomics, and I’ll suggest a more expansive approach that reveals CosMx SMI to be perhaps the most productive question-posing machine in molecular biology.
Data analysis begins with two fundamental pieces: a matrix of all cells’ gene expression, and their spatial locations:

As with any single cell dataset, we begin by using the gene expression matrix to call cell types. Then, by simply coloring our spatial map by cell type, we obtain an intricate picture of tissue structure.

Even this simplistic analysis is valuable: most experts examining these plots come away with new insights and new questions.
Questions answered with cell type + location:
– How is each immune cell type spread through this tumor? Where are the inflammatory vs. the suppressive macrophages trafficking?
– Do we see more memory T-cell invasion in post-treatment samples?
– What immune cell types tend to physically interact with each other?
Most early analyses of spatial transcriptomic data stop here. They wrap questions like the above in formal statistics (“spatial clustering” / “niche analysis”, “cell proximity analysis” and so on) and publish what is usually already a compelling story.
But if we take this analysis just a little farther, we can begin to ask a staggering number of questions. The power of CosMx isn’t its ability to get single cell expression profiles, nor its ability to describe spatial variation, it’s the ability to do both at the same time. By simultaneously measuring single cells’ phenotypes (gene expression) and environments (the phenotypes of surrounding cells), we can interrogate how phenotype responds to environment.
What can we say about a cell’s environment? Consider the below closeup of a PDAC (Pancreatic ductal adenocarcinoma) tumor. “T-cell 1” is in a lymphoid structure, surrounded by B-cells and endothelial cells. These cells are expressing certain levels of ligands that bind receptors on T-cells. In contrast, “T-cell 2” has invaded into the tumor bed and is mainly surrounded by tumor cells, plus a few macrophages, and these cells are signaling the T-cell with a different set of genes.

In short, we can easily derive over 1000 variables describing each cell’s environment. And now, for every cell type, we can measure how every gene responds to every environmental variable. This amounts to roughly the following:
20 cell types x 1000 single cell gene expression levels x 1000 environment gene expression levels = 20 million questions
Going even further, in a study across multiple tissues, we might ask these 20 million questions separately for each tissue. (Note that in a lower-plex technology, where a panel of ~300 genes might be devoted almost entirely to cell typing, the number of interesting questions to be asked drops precipitously.)
In short, a standard analysis of a single CosMx SMI run with our cloud-based informatics solution AtoMx Spatial Informatics Platform (SIP) can lead to millions of hypothesis tests. How many previously uncharacterized interactions might lie within that list? How many publications? That’s for you to find out.
Spatial Informatics Platform (SIP) can lead to millions of hypothesis tests. How many previously uncharacterized interactions might lie within that list? How many publications? That’s for you to find out.
Learn more today about how CosMx SMI and AtoMx SIP single-cell technology can further your research.
Questions on how phenotype responds to environment:
– How do tumor cells modulate gene expression in the face of T-cell attack?
– How do macrophages in the stroma differ from macrophages in the tumor interior?
– What genes do T-cells express when exposed to inflammatory cytokines?
This post may include information regarding CosMx SMI products for RNA detection, which products are not available in the member states of the European Unified Patent Court, as further described here.
SMI products for RNA detection, which products are not available in the member states of the European Unified Patent Court, as further described here.