Cell-cell interactions (CCIs) play critical roles in many biological processes such as cellular differentiation, tissue homeostasis, and immune response. With the rapid development of high throughput single-cell RNA sequencing (scRNA-seq) technologies, it is of high importance to identify CCIs from the ever-increasing scRNA-seq data. However, limited by the algorithmic constraints, current computational methods based on statistical strategies ignore some key latent information contained in scRNA-seq data with high sparsity and heterogeneity.
Researchers at the Harbin Institute of Technology have developed a deep learning framework named DeepCCI to identify meaningful CCIs from scRNA-seq data. Applications of DeepCCI to a wide range of publicly available datasets from diverse technologies and platforms demonstrate its ability to predict significant CCIs accurately and effectively. Powered by the flexible and easy-to-use software, DeepCCI can provide the one-stop solution to discover meaningful intercellular interactions and build CCI networks from scRNA-seq data.
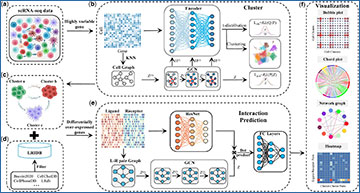
Workflow of the DeepCCI
(a) DeepCCI takes the scRNA-seq data as input. (b) DeepCCI clusters cells using the AE and the GCN jointly. (c) ScRNA-seq data with cell types. (d) LRIDB contains validated L–R interactions that were collected from several publicly literature-supported databases. (e) DeepCCI predicts the interactions between cell clusters using ResNet and GCN jointly. (f) DeepCCI offers several visualization outputs for different analytical tasks
Availability – The source code of DeepCCI is available online at https://github.com/JiangBioLab/DeepCCI
Yang W, Wang P, Luo M, Cai Y, Xu C, Xue G, Jin X, Cheng R, Que J, Pang F, Yang Y, Nie H, Jiang Q, Liu Z, Xu Z. (2023) DeepCCI: a deep learning framework for identifying cell-cell interactions from single-cell RNA sequencing data. Bioinformatics 39(10):btad596. [article]