Researchers at Dana-Farber Cancer Institute have created an AI-based tool that uses tumor gene sequencing data to predict the primary source of a patient’s cancer. The study, published in in Nature Medicine, suggests that this predictive tool, called OncoNPC, could help guide treatment of cancer and improve outcomes in difficult to diagnose cases.
The primary source of cancer is traditionally diagnosed by a standardized diagnostic work-up, including radiology and pathology assessments based on slides of cells taken from a tumor biopsy. In 3-5% of cancer cases, the original source of the tumor cannot be determined.
In these cases, patients are diagnosed with cancers of unknown primary (CUP) and have few treatment options because most treatments are approved for a specific type of cancer.
“This patient group has dismal outcomes,” says Dana-Farber researcher and senior author Alexander Gusev, PhD.
The team found that the AI model’s predictions could have value for these patients. A retrospective analysis suggested that this additional piece of diagnostic information about the primary source of the tumor could help doctors select treatments that improve survival.
“We see the OncoNPC prediction as a nudge, a way to provide a possible explanation for the cancer that helps point to appropriate treatment, including precision medicine,” says Gusev.
To build the model, the researchers trained and validated a machine learning classifier using the medical records of 36,445 patients with known primary tumors from three major cancer centers, including Dana-Farber. The records contained tumor genetic sequencing data and clinical information for each patient.
Gusev and first author Intae Moon, a graduate student at Massachusetts Institute of Technology and researcher in Gusev’s lab, chose to use a machine learning model that is interpretable, meaning that the reasoning behind the model’s prediction is more transparent than other forms of AI.
“We thought that this transparency would help clinicians trust the model,” says Moon. “It also could be clinically and biologically useful to see what genetic factors contributed to the model’s prediction, especially considering the enigmatic nature of CUP tumors.”
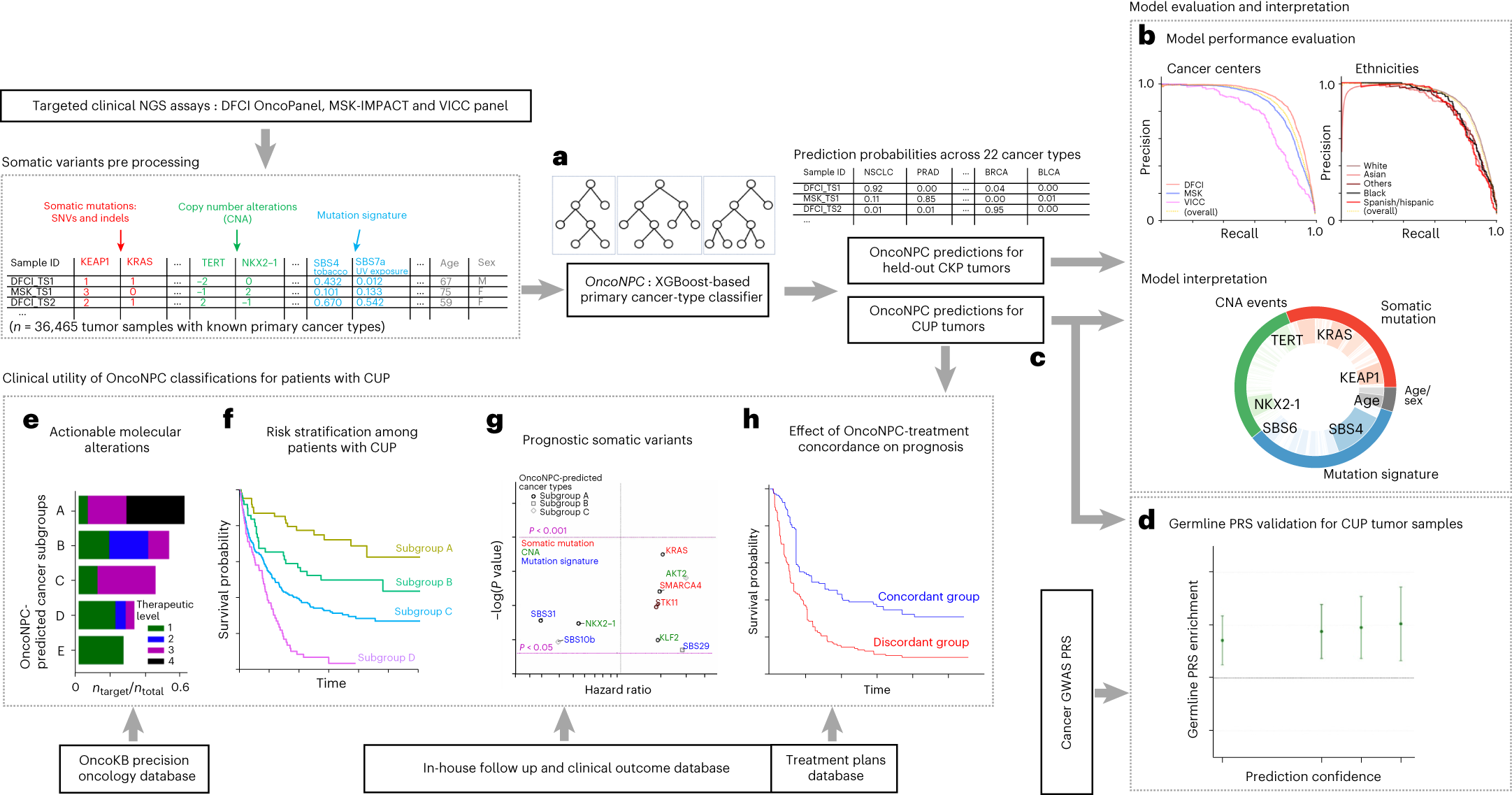
Overview of model development and analysis workflow
a, OncoNPC, an XGBoost-based classifier, was trained and evaluated using 36,465 cancer with known primary (CKP) tumor samples across 22 cancer types collected from three different cancer centers. b, OncoNPC performance was evaluated on the held-out tumor samples (n = 7,289). c, OncoNPC was applied to 971 CUP tumor samples at a single institution to predict primary cancer types. d–g, OncoNPC-predicted CUP subgroups were then investigated for association with elevated germline risk (d), actionable molecular alterations (e), overall survival (f) and prognostic somatic features (g). h, A subset of CUP patients with detailed treatment data was evaluated for treatment-specific outcomes.
OncoNPC, short for Oncology NGS-based Primary cancer type Classifier, accurately predicted the origin of about 80% of tumors with known types, including metastatic tumors, using a subset of cases that had not been used as training data. The model made high confidence predictions in 65% of the tumors, meaning it assessed its prediction as having a high probability of being correct. Those predictions were 95% accurate.
They then applied OncoNPC to a separate database of 971 CUP tumors from patients seen at Dana-Farber, where a team of experts had already made a substantial effort to identify the primary source of the tumor. OncoNPC was able to predict the tumor’s origin with high confidence for 400 out of 971 (41.2%) of the cases.
To validate these predictions, the team looked at inherited germline risks of cancer among these patients and found that the risks lined up with the predictions. Further, they looked at specific cases closely to determine if the data, including pathology results, patient history, and genetic mutations supported the prediction.
“Validation is a challenge because there is no ground truth. Existing methods failed to identify the origin,” says Gusev. “But the evidence we looked at showed us that the model is on the right track.”
To determine if an OncoNPC prediction might have value to patients, the team examined the outcomes of a subset of the patients with CUP. Patients who received treatments that matched the predicted primary tumor site had longer survival compared those receiving treatments that did not match the predictions. In addition, they found that the OncoNPC predictions would enable approximately 2.2 times as many CUP patients to be matched to approved targeted medicines.
“This could open the door to more precision treatment for these patients,” says Gusev.
The tool has so far been studied using retrospective data only. To determine if it could improve outcomes for patients, it would need to be tested in a clinical trial.
Gusev and Moon plan to build on OncoNPC by expanding the data it uses for prediction to include additional diagnostic information, such as pathology results.
In addition, they would like to collaborate with a community cancer center to learn more about how OncoNPC predictions could complement existing diagnostics. Cases of CUP might be more common in smaller cancer centers because they have fewer pathologists available to devote to difficult diagnoses.
“The appeal of this approach is that tumor panels are widely available and it’s easy to run those results through the algorithm to get a prediction,” says Gusev. “It could be valuable in settings with limited resources.”
Source – Dana-Farber Cancer Institute
Availability – https://github.com/itmoon7/onconpc
Moon I, LoPiccolo J, Baca SC, Sholl LM, Kehl KL, Hassett MJ, Liu D, Schrag D, Gusev A. (2023) Machine learning for genetics-based classification and treatment response prediction in cancer of unknown primary. Nat Med [Epub ahead of print]. [abstract]