Introduction to Polyadenylation
Polyadenylation is a critical process that occurs at the end of a gene’s transcription. This involves the addition of a polyA tail, a string of adenine nucleotides, to the RNA molecule. This tail is essential for RNA stability, transport, and translation into proteins. Most mammalian genes have multiple sites where this polyA tail can be added, known as polyA sites. The choice of these sites can result in different versions of RNA from the same gene, contributing to the diversity of the transcriptome, which is the complete set of RNA molecules in a cell.
The Role of CPA Machinery
The cleavage and polyadenylation (CPA) machinery is a collection of proteins that govern where and how the polyA tail is added to the RNA. Understanding how these proteins make their decisions is crucial for deciphering gene regulation and the resulting effects on cell function.
Introducing CPA-Perturb-seq
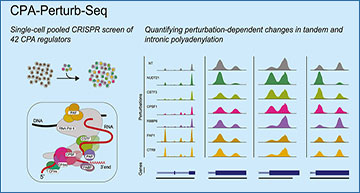
To explore the CPA machinery, scientists at the New York Genome Center have developed a new technique called CPA-Perturb-seq. This innovative method allows researchers to investigate the effects of disrupting various CPA proteins on polyA site choice across the entire transcriptome.
How CPA-Perturb-seq Works
CPA-Perturb-seq involves a few key steps:
- Multiplexed Perturbation Screening: Researchers target 42 different CPA regulators (proteins involved in the CPA process) and perturb, or disrupt, their function in a controlled manner.
- 3′ Single-Cell RNA Sequencing (scRNA-seq): This technique reads the RNA at its 3′ end (where the polyA tail is added) in individual cells, providing a detailed view of how each perturbation affects polyA site usage.
- Data Analysis Framework: The resulting data is analyzed to detect changes in polyadenylation patterns and to identify groups of co-regulated polyA sites.
Key Findings
Through CPA-Perturb-seq, several significant discoveries were made:
- Intronic PolyA Sites: These are polyA sites located within the introns (non-coding regions) of genes. Researchers found that specific components of the nuclear RNA life cycle, including elongation, splicing, termination, and surveillance, regulate these sites.
- Regulatory Modules: They identified modules of polyA sites that are co-regulated, meaning they respond similarly to perturbations in CPA machinery.
Deep Neural Network – APARENT-Perturb
To further understand how different polyA sites respond to perturbations, researchers trained a deep neural network called APARENT-Perturb. This model predicts how changes in the regulatory environment affect polyA site usage by analyzing the sequence and structure around these sites. It helps in understanding the complex interactions between different regulatory proteins and their impact on gene expression.
Implications for Post-Transcriptional Regulation
The findings from CPA-Perturb-seq emphasize the complexity and precision of post-transcriptional regulation, which is how cells control the fate of RNA molecules after they are transcribed from DNA. By using advanced techniques like single-cell sequencing and deep learning, scientists can uncover the nuanced mechanisms that govern gene expression.
Conclusion
The development of CPA-Perturb-seq and the insights gained from it represent a significant advancement in the field of genetics. By understanding how CPA machinery regulates polyA site choice, researchers can better comprehend the layers of gene regulation that contribute to cellular function and diversity. This knowledge has the potential to inform the development of new therapeutic strategies for diseases where gene expression is disrupted.
CPA-Perturb-seq is a powerful tool that sheds light on the regulation of polyadenylation, highlighting the intricate control mechanisms that maintain proper gene function in mammalian cells.
Kowalski MH, Wessels HH, Linder J, Dalgarno C, Mascio I, Choudhary S, Hartman A, Hao Y, Kundaje A, Satija R. (2024) Multiplexed single-cell characterization of alternative polyadenylation regulators. Cell [Epub ahead of print]. [abstract]