The advent of single-cell RNA-sequencing (scRNA-seq) has driven significant computational methods development for all steps in the scRNA-seq data analysis pipeline, including filtering, normalization, and clustering. The large number of methods and their resulting parameter combinations has created a combinatorial set of possible pipelines to analyze scRNA-seq data, which leads to the obvious question: which is best? Several benchmarking studies have sought to compare methods to answer this, but frequently find variable performance depending on dataset and pipeline characteristics. Alternatively, the large number of publicly available scRNA-seq datasets along with advances in supervised machine learning raise a tantalizing possibility: could the optimal pipeline be predicted for a given dataset?
Researchers at the Lunenfeld-Tanenbaum Research Institute begin to answer this question by applying 288 scRNA-seq analysis pipelines to 86 datasets and quantifying pipeline success via a range of measures evaluating cluster purity and biological plausibility. The researchers build supervised machine learning models to predict pipeline success given a range of dataset and pipeline characteristics. They find both that prediction performance is significantly better than random and that in many cases pipelines predicted to perform well provide clustering outputs similar to expert-annotated cell type labels. Finally, the researchers identify characteristics of scRNA-seq datasets that correlate with strong prediction performance that could guide when such prediction models may be useful.
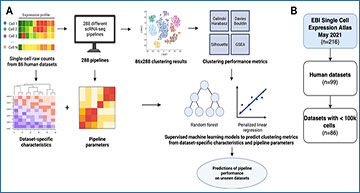
A Overview of the machine learning workflow: 288 clustering pipelines were run over each dataset and the success of each was quantified with 4 unsupervised metrics. Dataset– and pipeline-specific features were then computed and given as input to supervised machine learning models to predict metric values. B 86 human datasets in the EBI Single Cell Expression Atlas containing <100k cells as of May 2021 were selected for this study. C Characteristics of the 86 datasets used as input to predictive models of dataset-specific pipeline performance. These include median values of metrics frequently used to quality control at the cell level (e.g., percentage of mitochondrial counts) as well as principal components of average expression values per dataset jointly decomposed. Each characteristic was scaled in the training set to follow a standard normal distribution. The means and variances before scaling of each characteristic in the training set were used to scale the corresponding characteristics in the test set to prevent train-test leakage.
Fang C, Selega A, Campbell KR. (2023) Beyond benchmarking: towards predictive models of dataset-specific single-cell RNA-seq pipeline performance. bioRXiv [online preprint]. [article]