Single-cell datasets are routinely collected to investigate changes in cellular state between control cells and the corresponding cells in a treatment condition, such as exposure to a drug or infection by a pathogen. To better understand heterogeneity in treatment response, it is desirable to deconvolve variations enriched in treated cells from those shared with controls. However, standard computational models of single-cell data are not designed to explicitly separate these variations.
Researchers at the University of Washington have developed contrastive variational inference (contrastiveVI), a framework for deconvolving variations in treatment–control single-cell RNA sequencing (scRNA-seq) datasets into shared and treatment-specific latent variables. Using three treatment–control scRNA-seq datasets, the researchers apply contrastiveVI to perform a variety of analysis tasks, including visualization, clustering and differential expression testing. They found that contrastiveVI consistently achieves results that agree with known ground truths and often highlights subtle phenomena that may be difficult to ascertain with standard workflows. The researchers conclude by generalizing contrastiveVI to accommodate joint transcriptome and surface protein measurements.
Overview of contrastiveVI
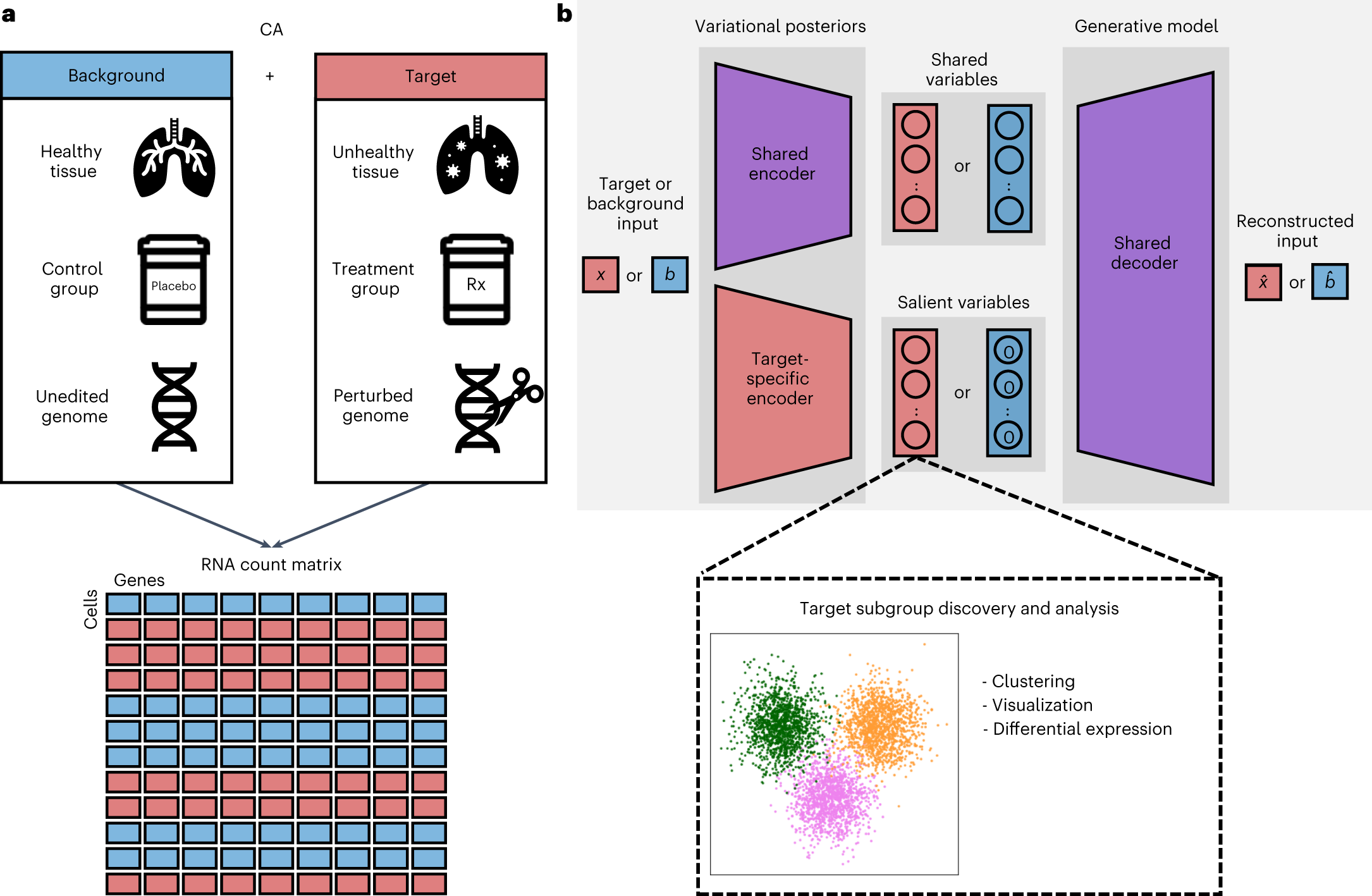
For a target dataset of interest and the corresponding background dataset, contrastiveVI separates the variations shared between the two datasets and the variations enriched in the target dataset. a, Example background and target data pairs. Samples from both conditions produce an RNA count matrix with each cell labeled as background or target. Rx, prescription. CA, contrastive analysis. b, Schematic of the contrastiveVI model. A shared encoder network embeds a cell, whether target (red) or background (blue), into the model’s shared latent space, which captures variations common to target and background cells. A second target cell-specific encoder embeds target cells into the model’s salient latent space, which captures variations enriched in the target data and not present in the background. For background cells, the values of the salient latent factors are fixed to be a zero vector. Both target and background cells’ latent representations are transformed back to the original gene expression space using a single shared decoder network.
Availability – https://github.com/suinleelab/contrastiveVI
Weinberger E, Lin C, Lee SI. (2023) Isolating salient variations of interest in single-cell data with contrastiveVI. Nat Methods [Epub ahead of print]. [abstract]