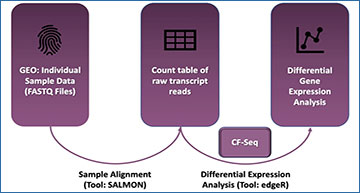
Researchers studying cystic fibrosis (CF) pathogens have produced numerous RNA-seq datasets which are available in the gene expression omnibus (GEO). Although these studies are publicly available, substantial computational expertise and manual effort are required to compare similar studies, visualize gene expression patterns within studies, and use published data to generate new experimental hypotheses. Furthermore, it is difficult to filter available studies by domain-relevant attributes such as strain, treatment, or media, or for a researcher to assess how a specific gene responds to various experimental conditions across studies.
To reduce these barriers to data re-analysis, Dartmouth College researchers have developed an R Shiny application called CF-Seq, which works with a compendium of 128 studies and 1,322 individual samples from 13 clinically relevant CF pathogens. The application allows users to filter studies by experimental factors and to view complex differential gene expression analyses at the click of a button. These researchers present a series of use cases that demonstrate the application is a useful and efficient tool for new hypothesis generation.
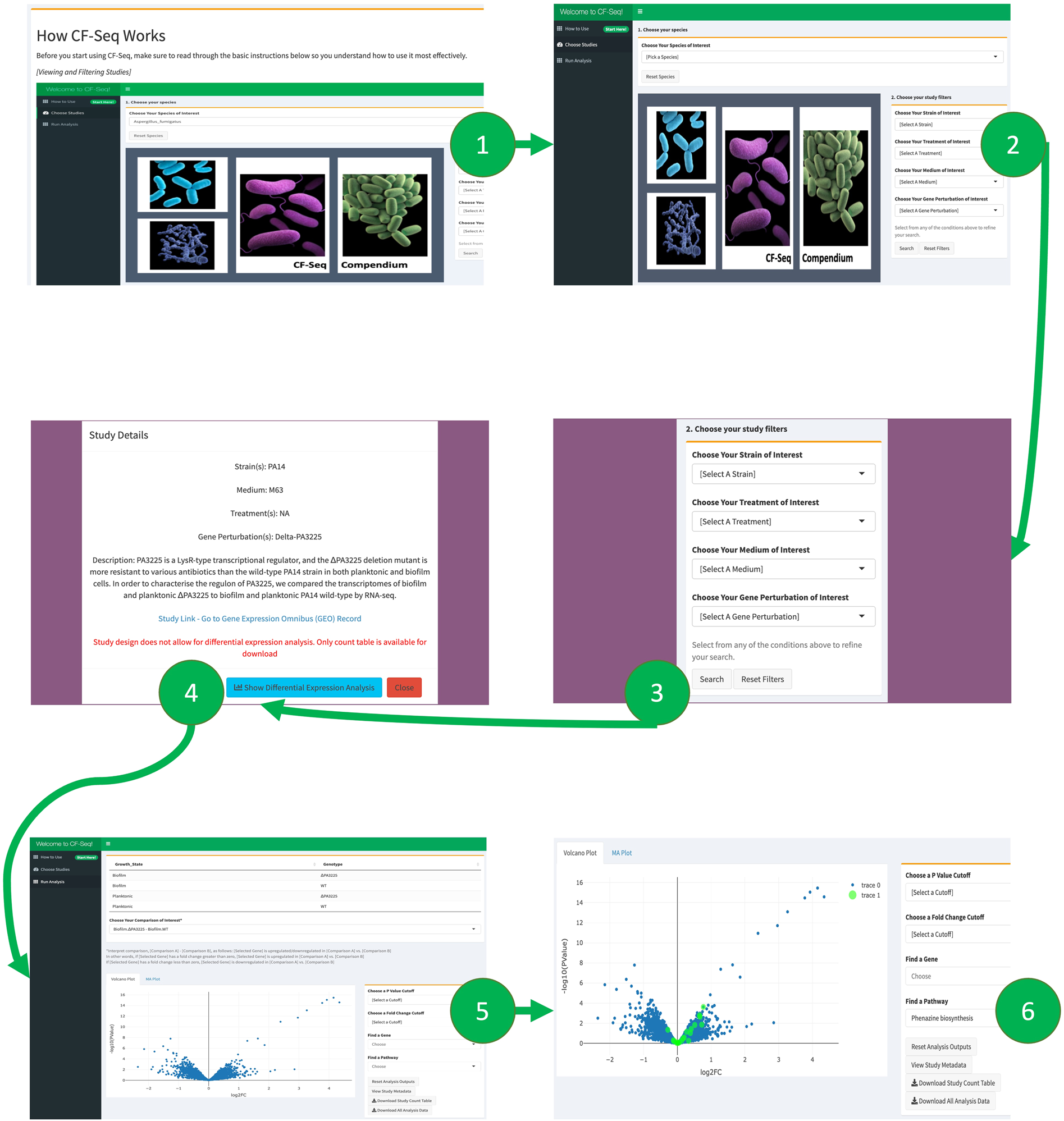
Application workflow for CF-Seq users
Panel 1 shows the starting window of the application, where users are presented with a manual that explains the functionality and purpose of the application. Users are then directed to the study view screen, shown in panel 2, where they can select a species of interest and view available RNA-Seq studies. Panel 3 shows how filters can be applied to delineate studies with certain experimental characteristics (strain, media, treatment, gene perturbed). Panel 4 offers a look at the metadata that can be examined for each individual study. Panels 5 and 6 show the study analysis window, where analysis tables and figures can be generated for all experimental comparisons, individual genes may be highlighted, P value and fold change cutoffs can be selected, and differentially expressed genes on selected KEGG pathways can be highlighted when KEGG pathway information is available (Panel 6). For certain studies, users can also highlight other biological features, such as GO terms, COG categories, and functional descriptions of genes (e.g., “serine/threonine protein kinase”)
Availability – CF-Seq is available at: http://scangeo.dartmouth.edu/CFSeq/
Neff SL, Hampton TH, Puerner C et al. (2022) CF-Seq, an accessible web application for rapid re-analysis of cystic fibrosis pathogen RNA sequencing studies. Sci Data 9, 343. [article]