Transposable elements (TEs) serve as both insertional mutagens and regulatory elements in cells, and their aberrant activity is increasingly being revealed to contribute to diseases and cancers. However, measuring the transcriptional consequences of nonreference and young TEs at individual loci remains challenging with current methods, primarily due to technical limitations, including short read lengths generated and insufficient coverage in target regions.
Researchers at Sichuan University have developed a long-read targeted RNA sequencing method, Cas9-assisted profiling TE expression sequencing (capTEs), for quantitative analysis of transcriptional outputs for individual TEs, including transcribed nonreference insertions, noncanonical transcripts from various transcription patterns and their correlations with expression changes in related genes. This method selectively identified TE-containing transcripts and outputted data with up to 90% TE reads, maintaining a comparable data yield to whole-transcriptome sequencing. The researchers applied capTEs to human cancer cells and found that internal and inserted Alu elements may employ distinct regulatory mechanisms to upregulate gene expression. They expect that capTEs will be a critical tool for advancing our understanding of the biological functions of individual TEs at the locus level, revealing their roles as both mutagens and regulators in biological and pathogenic processes.
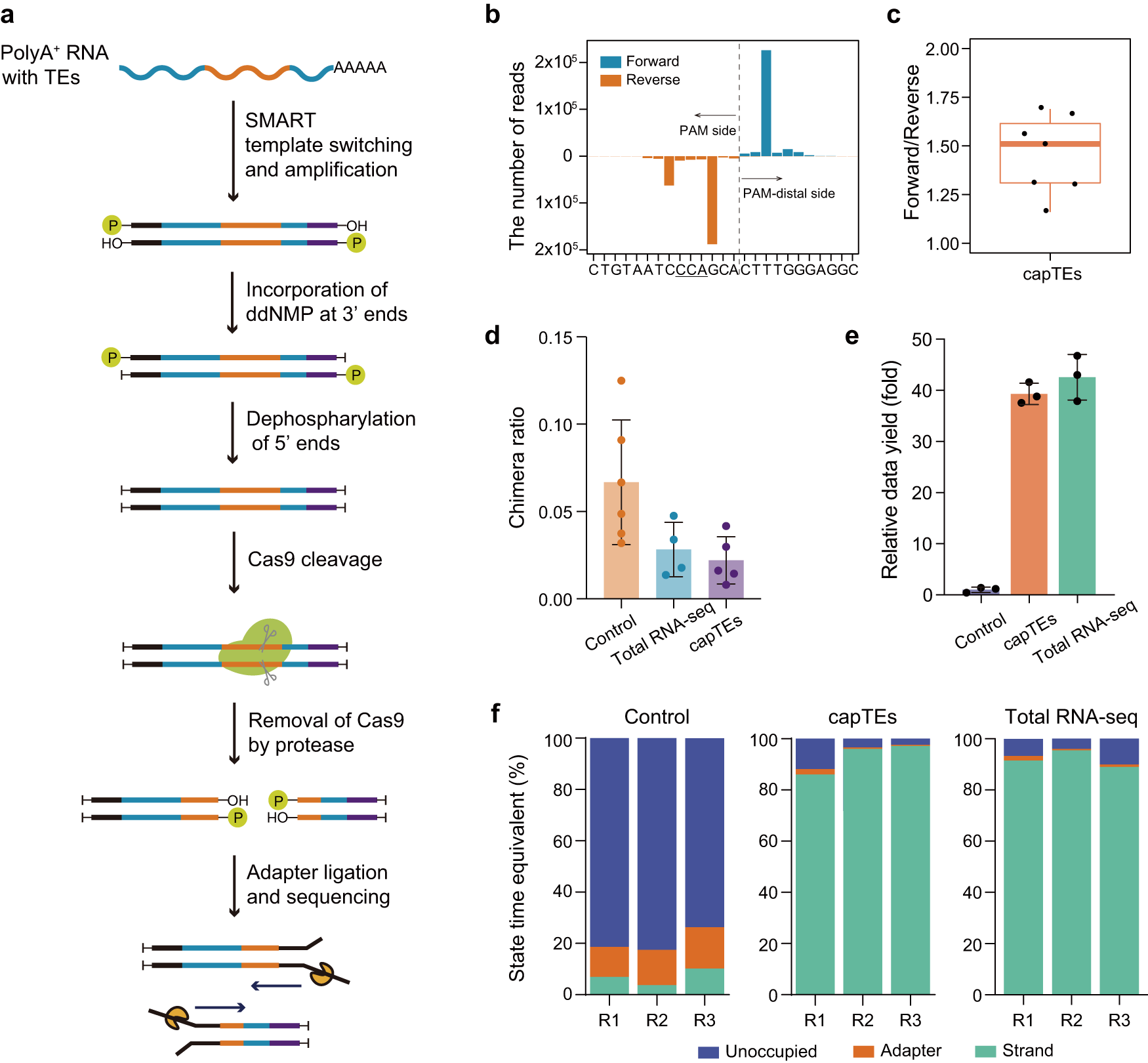
Overview of capTEs
a Schematic of the experimental workflow. The full-length ds-cDNA library was constructed from total RNA usng SMART technology, and cDNA ends were inactivated by ddGMP incorporation to block 3’ hydroxyl residues and dephosphorylation to remove 5’ phosphate residues. Then, new DNA ends were created by Cas9-gRNAs targeting specific sequences. After the release of Cas9 from the cleavage sites by the thermolabile protease, sequencing adapters were ligated to the cleavage sites for subsequent sequencing. b Histogram displaying the strand distribution of capTEs data in the targeted region of Alu gRNA. The x-axis shows the position where the read starts or ends. The underlined uppercase letters represent the PAM sequence, and the dashed line marks the Cas9 cut site. c Boxplot showing the strand ratio of capTEs data (n = 7). The box edges and whiskers indicate the median, upper and lower quartiles (the 25th and 75th percentiles) and 1.5 × interquartile range, respectively. d Bar plots showing side reaction rates of control (orange, n = 6), total RNA-seq (blue, n = 4) and capTEs (purple, n = 5). The side reaction rate is determined by the fraction of hybrid reads among all reads containing spike-in sequences. Error bars represent standard deviation. e Bar plot showing the data outputs of capTEs (orange, n = 3) and total RNA-seq (green, n = 3) relative to the control (gray–purple, n = 3), where the control is normalized to 1. Error bars represent standard deviation. f Stacked bar plots showing the state composition of available pores in control, capTEs and total RNA-seq: unoccupied pores (blue), adapter-occupied pores (orange) and DNA strand-occupied pores (green). The proportion (y-axis) is determined by the occupied time. d–f In the control, nCATS is directly applied to capture TE transcripts.
Availability – The source code is available at: https://github.com/KeyingLu/capTEs.
Li X, Lu K, Chen X, Tu K, Xie D. (2023) capTEs enables locus-specific dissection of transcriptional outputs from reference and nonreference transposable elements. Commun Biol 6(1):974. [article]