Precise mechanism-based gene expression signatures (GES) have been developed in appropriate in vitro and in vivo model systems, to identify important cancer-related signaling processes. However, some GESs originally developed to represent specific disease processes, primarily with an epithelial cell focus, are being applied to heterogeneous tumor samples where the expression of the genes in the signature may no longer be epithelial-specific. Therefore, unknowingly, even small changes in tumor stroma percentage can directly influence GESs, undermining the intended mechanistic signaling.
Using colorectal cancer as an exemplar, researchers at Queen’s University Belfast deployed numerous orthogonal profiling methodologies, including laser capture microdissection, flow cytometry, bulk and multiregional biopsy clinical samples, single-cell RNA sequencing and finally spatial transcriptomics, to perform a comprehensive assessment of the potential for the most widely used GESs to be influenced, or confounded, by stromal content in tumor tissue. To complement this work, the researchers generated a freely-available resource, ConfoundR; https://confoundr.qub.ac.uk/, that enables users to test the extent of stromal influence on an unlimited number of the genes/signatures simultaneously across colorectal, breast, pancreatic, ovarian and prostate cancer datasets.
Findings presented here demonstrate the clear potential for misinterpretation of the meaning of GESs, due to widespread stromal influences, which in-turn can undermine faithful alignment between clinical samples and preclinical data/models, particularly cell lines and organoids, or tumor models not fully recapitulating the stromal and immune microenvironment.
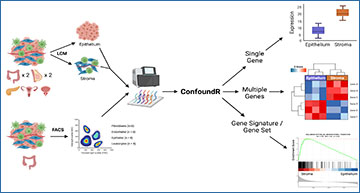
The ConfoundR resource enables stromal influence estimation in cancer tissue
A, Schematic overview of the cohorts and analyses available within the ConfoundR app, accessible via https://confoundr.qub.ac.uk/. B, Expression Boxplots analysis module of ConfoundR enabling the expression of a single gene to be compared between stroma and epithelium samples in each of the ConfoundR datasets. C, Expression Heatmap analysis module of ConfoundR enabling the expression of multiple genes to be visually compared between stroma and epithelium samples in each of the ConfoundR datasets. D, GSEA analysis module of ConfoundR allowing GSEA of existing gene sets from established gene set collections or custom user defined gene sets to be performed comparing stroma with epithelium in each of the ConfoundR datasets.
Efforts to faithfully align preclinical models of disease using phenotypically-designed GESs must ensure that the signatures themselves remain representative of the same biology when applied to clinical samples.
Fisher NC, Byrne RM, Leslie H, Wood C, Legrini A, Cameron AJ, Ahmaderaghi B, Corry SM, Malla SB, Amirkhah R, McCooey AJ, Rogan E, Redmond KL, Sakhnevych S, Domingo E, Jackson J, Loughrey MB, Leedham S, Maughan T, Lawler M, Sansom OJ, Lamrock F, Koelzer VH, Jamieson NB, Dunne PD. (2022) Biological Misinterpretation of Transcriptional Signatures in Tumor Samples Can Unknowingly Undermine Mechanistic Understanding and Faithful Alignment with Preclinical Data. Clin Cancer Res 28(18):4056-4069. [article]