The growing number of available single-cell gene expression datasets from different species creates opportunities to explore evolutionary relationships between cell types across species. Cross-species integration of single-cell RNA-sequencing data has been particularly informative in this context. However, in order to do so robustly it is essential to have rigorous benchmarking and appropriate guidelines to ensure that integration results truly reflect biology.
EMBL-EBI researchers benchmark 28 combinations of gene homology mapping methods and data integration algorithms in a variety of biological settings. They examine the capability of each strategy to perform species-mixing of known homologous cell types and to preserve biological heterogeneity using 9 established metrics. They also develop a new biology conservation metric to address the maintenance of cell type distinguishability. Overall, scANVI, scVI and SeuratV4 methods achieve a balance between species-mixing and biology conservation. For evolutionarily distant species, including in-paralogs is beneficial. SAMap outperforms when integrating whole-body atlases between species with challenging gene homology annotation. The researchers provide their freely available cross-species integration and assessment pipeline to help analyse new data and develop new algorithms.
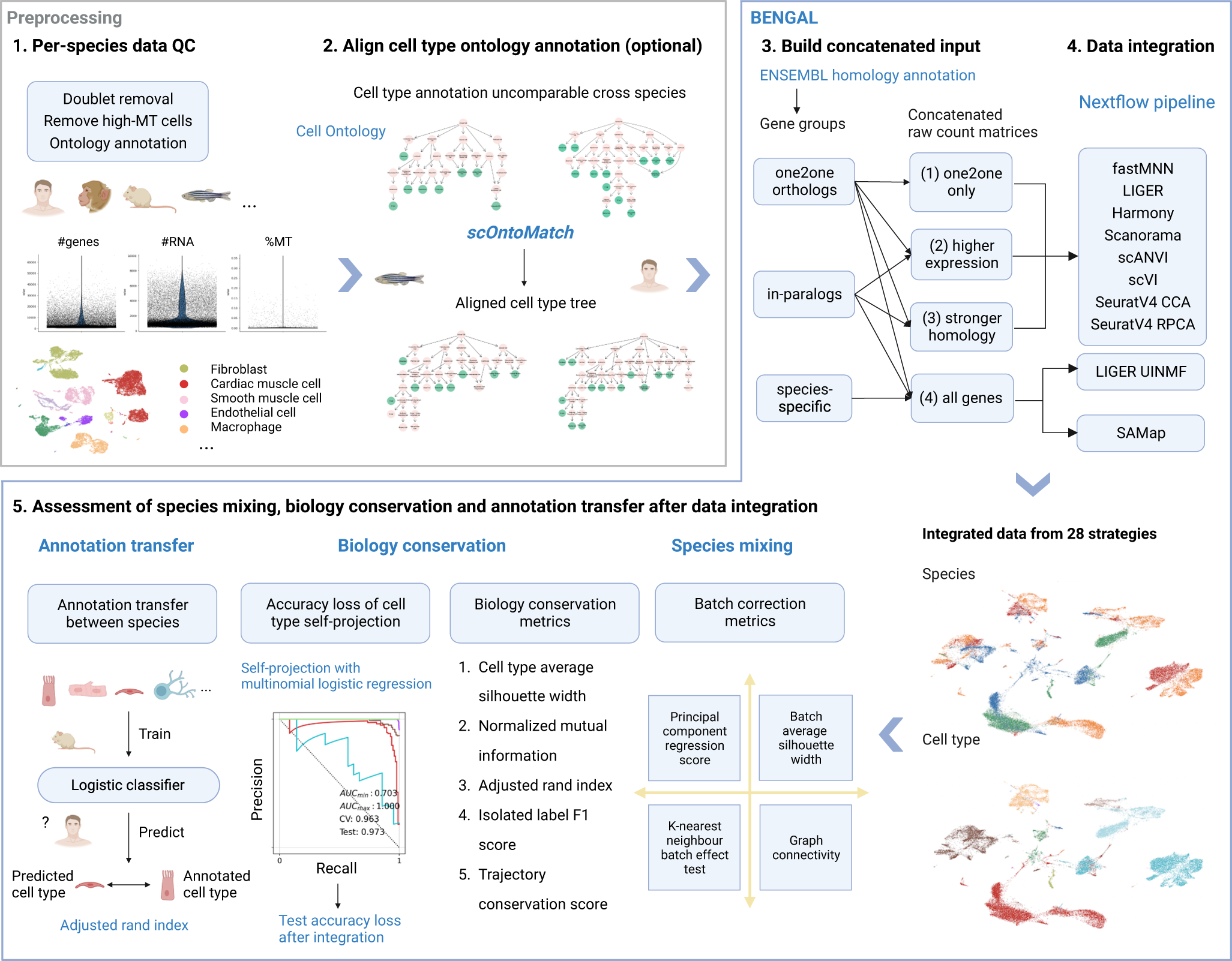
Schematic of the BENGAL pipeline
1 Quality control of input data is performed prior to data integration. and is not part of BENGAL. Potential doublets and low-quality cells expressing high mitochondrial genes should be removed. Cell ontology annotations are collected from atlases or data portals or curated from the originally published annotation. 2 When the granularity of ontology annotation is incomparable across datasets, one-to-one homology between cell types needs to be robustly aligned. We developed scOntoMatch to find the appropriate annotation granularity and align cell type hierarchies across given datasets (see Methods). 3 Genes are grouped and translated across species by homology defined in ENSEMBL multiple species comparison tool. Raw count matrices are then concatenated across species using four possible homology matching methods respective to method inputs. 4 Run 9 integration algorithms to generate integrated output. 5 Perform integration assessment from species mixing, biology conservation and cell type annotation transfer. BENGAL, BENchmarking strateGies for cross-species integrAtion of singLe-cell RNA sequencing data; QC, quality control; MT, mitochondrial; SCCAF, single cell clustering assessment framework; AUC, area under the curve; CV, cross-validation.
Availability -The BENGAL pipeline is available at https://github.com/Functional-Genomics/BENGAL and the version of code used in this study is available via Zenodo with https://doi.org/10.5281/zenodo.8268784
Song Y, Miao Z, Brazma A, Papatheodorou I. (2023) Benchmarking strategies for cross-species integration of single-cell RNA sequencing data. Nat Commun 14(1):6495. [article]