In silico cell-type deconvolution from bulk transcriptomics data is a powerful technique to gain insights into the cellular composition of complex tissues. While first-generation methods used precomputed expression signatures covering limited cell types and tissues, second-generation tools use single-cell RNA sequencing data to build custom signatures for deconvoluting arbitrary cell types, tissues, and organisms. This flexibility poses significant challenges in assessing their deconvolution performance.
Researchers at the University of Innsbruck comprehensively benchmark second-generation tools, disentangling different sources of variation and bias using a diverse panel of real and simulated data. This study highlights the strengths, limitations, and complementarity of state-of-the-art tools shedding light on how different data characteristics and confounders impact deconvolution performance. The researchers provide the scientific community with an ecosystem of tools and resources, omnideconv, simplifying the application, benchmarking, and optimization of deconvolution methods.
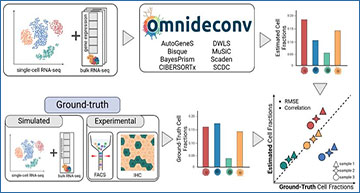
omnideconv ecosystem and benchmark
(A) The omnideconv benchmarking ecosystem offers five tools (from left to right): the R package omnideconv providing a unified interface to deconvolution methods, the pseudo-bulk simulation method SimBu, the deconvData data repository, the deconvBench benchmarking pipeline in Nextflow and the web-app deconvExplorer. (B) Outline of the benchmark experiment: scRNA-seq and bulk RNA-seq data is used as input for several methods, and a unified output of estimated cell-type fractions for each bulk sample is calculated. We compare the estimated fractions to ground-truth fractions (known from pseudo-bulks or FACS/IHC experiments) and compute performance measures per method and cell type. (C) Several challenges in cell-type deconvolution are addressed in this benchmark: (1) cell types show total mRNA bias; (2) scRNA-seq datasets vary by technology, tissue and disease; (3) a fraction of the cells may be of unknown type, since the scRNA-seq reference does not necessarily contain all cell types present in the bulk mixture; (4) some cell types are more similar on a transcriptomic level, leading to “spillover” towards similar cell types. (D) We evaluated two major parameters that can often be adapted by users of deconvolution methods and affect estimation quality: (1) the number of cells for each annotated cell type in the scRNA-seq reference dataset and (2) the level of annotation granularity. This figure was created with Biorender.com.
Availability – https://omnideconv.org/
Dietrich A, Merotto L, Pelz K, Eder B, Zackl C, Reinisch K, F, Marini F, Sturm G, List M, Finotello F. (2024) Benchmarking second-generation methods for cell-type deconvolution of transcriptomic data. bioRXiv [Epub ahead of print]. [article]