Single-cell RNA-sequencing (scRNA-seq) enables researchers to quantify transcriptomes of thousands of cells simultaneously and study transcriptomic changes between cells. scRNA-seq datasets increasingly include multisubject, multicondition experiments to investigate cell-type-specific differential states (DS) between conditions. This can be performed by first identifying the cell types in all the subjects and then by performing a DS analysis between the conditions within each cell type. Naïve single-cell DS analysis methods that treat cells statistically independent are subject to false positives in the presence of variation between biological replicates, an issue known as the pseudoreplicate bias. While several methods have already been introduced to carry out the statistical testing in multisubject scRNA-seq analysis, comparisons that include all these methods are currently lacking.
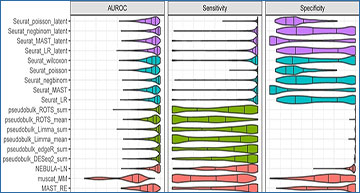
University of Turku researchers performed a comprehensive comparison of 18 methods for the identification of DS changes between conditions from multisubject scRNA-seq data. Their results suggest that the pseudobulk methods performed generally best. Both pseudobulks and mixed models that model the subjects as a random effect were superior compared with the naïve single-cell methods that do not model the subjects in any way. While the naïve models achieved higher sensitivity than the pseudobulk methods and the mixed models, they were subject to a high number of false positives. In addition, accounting for subjects through latent variable modeling did not improve the performance of the naïve methods.
Results of the simulation based on a reference-free negative binomial generative model. Each boxplot shows values for 1280 simulated datasets with varying data properties.
Junttila S, Smolander J, Elo LL. (2022) Benchmarking methods for detecting differential states between conditions from multi-subject single-cell RNA-seq data. Brief Bioinform [Epub ahead of print]. [article]