Single cell mRNA sequencing technologies have transformed our understanding of cellular heterogeneity and identity. For sensitive discovery or clinical marker estimation where high transcript capture per cell is needed only plate-based techniques currently offer sufficient resolution.
University of Copenhagen researchers present a performance evaluation of four different plate-based scRNA-seq protocols. Their evaluation is aimed towards applications taxing high gene detection sensitivity, reproducibility between samples, and minimum hands-on time, as is required, for example, in clinical use. The researchers included two commercial kits, NEBNext® Single Cell/ Low Input RNA Library Prep Kit (NEB®), SMART-seq® HT kit (Takara®), and the non-commercial protocols Genome & Transcriptome sequencing (G&T) and SMART-seq3 (SS3). G&T delivered the highest detection of genes per single cell. SS3 presented the highest gene detection per single cell at the lowest price. Takara® kit presented similar high gene detection per single cell, and high reproducibility between samples, but at the absolute highest price. NEB® delivered a lower detection of genes but remains an alternative to more expensive commercial kits.
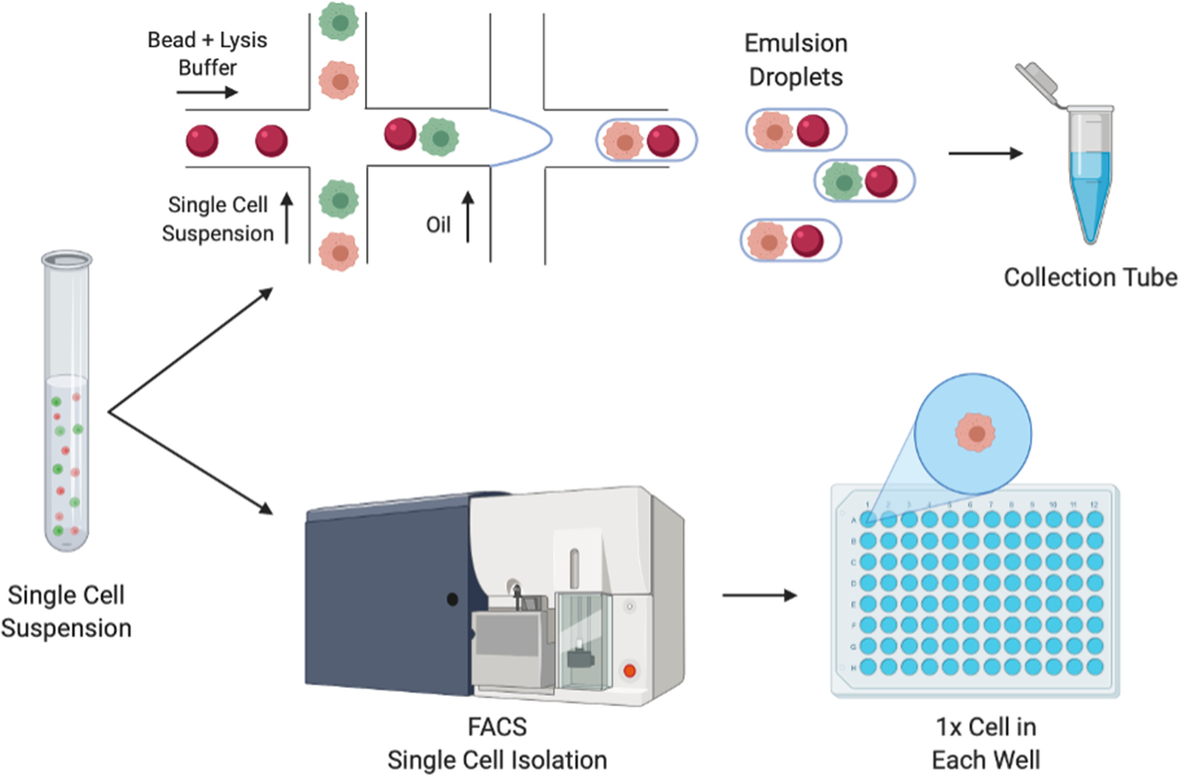
Workflow for droplet (top) and plate-based (bottom) single cell sequencing technologies
Droplet-based approaches combine primer-covered beads and single cells in emulsion droplets. In the droplets the single cell is lysed and reverse transcription (RT) is performed. Following RT, the synthesised cDNA is pooled and processed in bulk. In the Plate-based approaches, single cells are deposited in lysis-buffer filled chambers of a PCR plate or tube. Fluorescent activated cell sorting (FACS) may be used for this isolation of single cells
For the tested kits the researchers found that ease-of-use came at higher prices. Takara can be selected for its ease-of-use to analyse a few samples, but they recommend the cheaper G&T-seq or SS3 for laboratories where a substantial sample flow can be expected.
Probst V, Simonyan A, Pacheco F, Guo Y, Nielsen FC, Bagger FO. (2022) Benchmarking full-length transcript single cell mRNA sequencing protocols. BMC Genomics 23(1):860. [article]