Simulation of RNA-seq reads is critical in the assessment, comparison, benchmarking, and development of bioinformatics tools. Yet the field of RNA-seq simulators has progressed little in the last decade. To address this need University of Pennsylvania researchers have developed BEERS2, which combines a flexible and highly configurable design with detailed simulation of the entire library preparation and sequencing pipeline. BEERS2 takes input transcripts (typically fully-length mRNA transcripts with polyA tails) from either customizable input or from CAMPAREE simulated RNA samples. It produces realistic reads of these transcripts as FASTQ, SAM, or BAM formats with the SAM or BAM formats containing the true alignment to the reference genome. It also produces true transcript-level quantification values. BEERS2 combines a flexible and highly configurable design with detailed simulation of the entire library preparation and sequencing pipeline and is designed to include the effects of polyA selection and RiboZero for ribosomal depletion, hexamer priming sequence biases, GC-content biases in PCR amplification, barcode read errors, and errors during PCR amplification. These characteristics combine to make BEERS2 the most complete simulation of RNA-seq to date. Finally, the researchers demonstrate the use of BEERS2 by measuring the effect of several settings on the popular Salmon pseudoalignment algorithm.
BEERS2 Overview
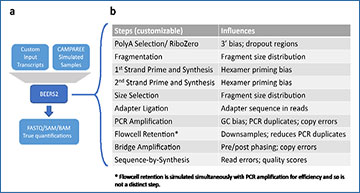
(a) BEERS2 is a customizable pipeline which processes input transcripts (typically fully-length mRNA transcripts with polyA tails) from either customizable input or from CAMPAREE simulated RNA samples. It produces realistic reads of these transcripts as FASTQ, SAM, or BAM formats with the SAM or BAM formats containing the true alignment to the reference genome. It also produces true transcript-level quantification values. (b) The default pipeline simulates the Illumina Stranded mRNA library preparation process, followed by sequencing. Each step influences one or more aspects of the output sample through configurable parameters.
Availability– The code for the BEERS2 simulator is available under an open-source GPLv3 license at https://github.com/itmat/BEERS2.
Brooks TG, Lahens NF, Mrcela A, Sarantapoulou D, Nayak S, Naik A, Sengupta S, Choi PS, Grant GR. (2023) BEERS2: RNA-Seq simulation through high fidelity in silico modeling. bioRXiv [online preprint]. [abstract]