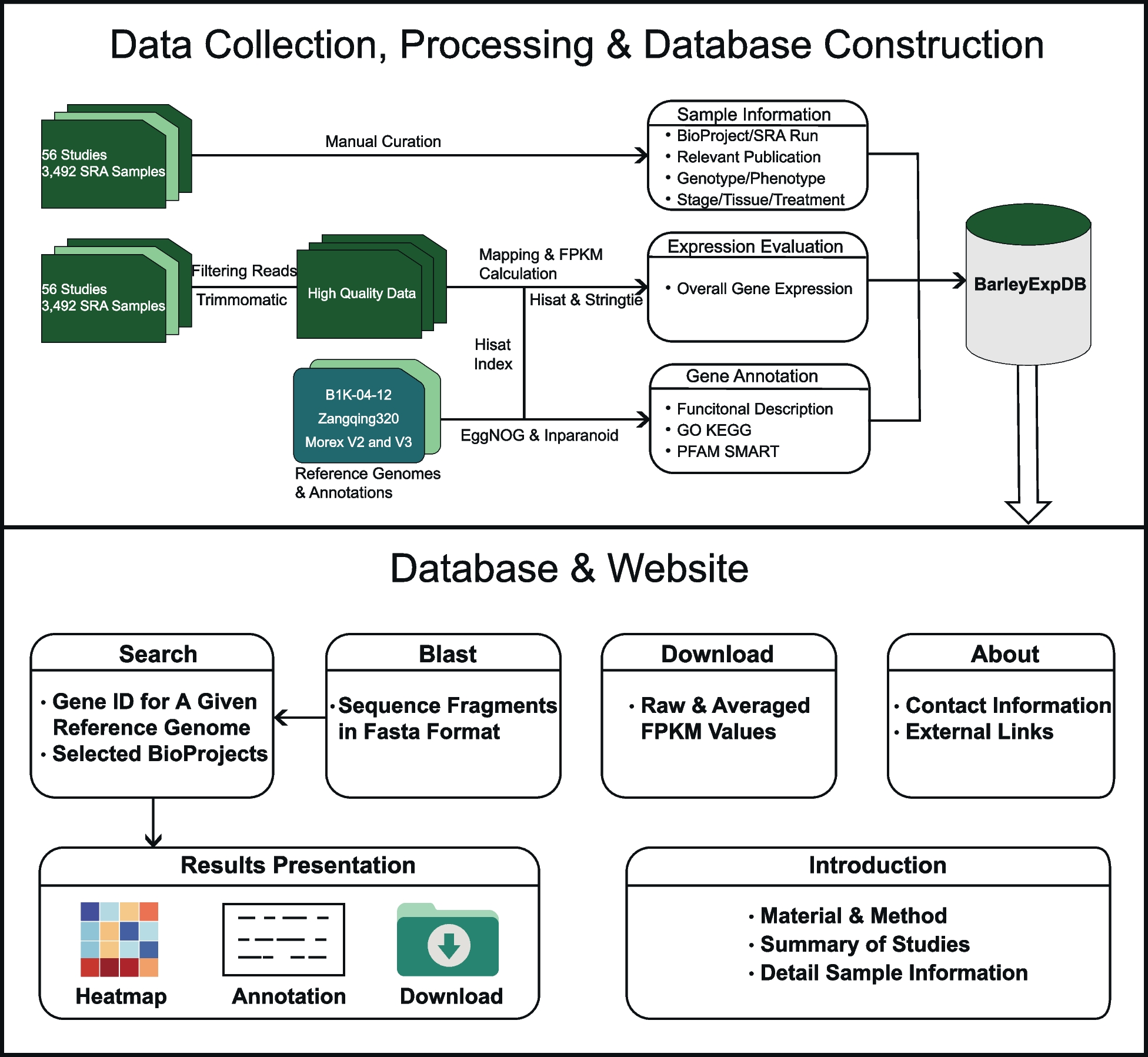
RNA-sequencing (RNA-seq) has been widely used to study the dynamic expression patterns of transcribed genes, which can lead to new biological insights. However, processing and analyzing these huge amounts of histological data remains a great challenge for wet labs and field researchers who lack bioinformatics experience and computational resources.
Researchers at Jiangxi Agricultural University have developed BarleyExpDB, an easy-to-operate, free, and web-accessible database that integrates transcriptional profiles of barley at different growth and developmental stages, tissues, and stress conditions, as well as differential expression of mutants and populations to build a platform for barley expression and visualization. The expression of a gene of interest can be easily queried by searching by known gene ID or sequence similarity. Expression data can be displayed as a heat map, along with functional descriptions as well as Gene Ontology, Kyoto Encyclopedia of Genes and Genomes, Proteins Families Database, and Simple Modular Architecture Research Tool annotations.
Workflow of BarleyExpDB
BarleyExpDB will serve as a valuable resource for the barley research community to leverage the vast publicly available RNA-seq datasets for functional genomics research and crop molecular breeding.
Availability – http://barleyexp.com/
Li T, Li Y, Shangguan H, Bian J, Luo R, Tian Y, Li Z, Nie X, Cui L. (2023 BarleyExpDB: an integrative gene expression database for barley. BMC Plant Biol 23(1):170. [article]