It is a revolutionary new way of looking at cells that is already having an impact across medical science. Now, the close-up lens of spatial transcriptomics is being turned to plants.
The cutting-edge approach allows you to measure the activity of genes in a tissue or cell and use imaging to visually map out where the activity is taking place.
Researchers at the Earlham Institute are using spatial transcriptomics to reveal insights into how individual plant cells are organised – in what is thought to be first-of-its-kind work in the UK.
Scientists have found themselves between a rock and a hard place when choosing a sequencing method to study the role of RNA in the normal functioning of a tissue.
Traditional bulk RNA sequencing loses differences between individual cells, while single-cell RNA sequencing lacks contextual information about the cell’s surroundings, losing important data about positioning and communication within a tissue.
Now, the emerging field of spatial transcriptomics means it is possible to visualise many RNA molecules in cells while retaining more of this detailed information. This method of mapping the location of gene activity in tissue samples uses imaging to mark which RNA species occur in a cell – and where they cluster.
The technique has already provided insights and deductions about gene function in developmental biology, cancer research, immunology, and neuroscience. But, while it has been used to study mammalian cells, there are only a handful of studies where it is applied to plants.
Ashleigh Lister, a Senior Research Assistant in Dr Iain Macaulay’s group at the Earlham Institute, is one of the first people in the UK to study single-cell and spatial transcriptomics in the plant kingdom.
Lister has been working with researchers across Norwich Research Park on transcription in wheat pollen, grain, and the basal section of wheat spikes. She describes a project that is both exciting and challenging.
“This type of work has never been done in plants and there is no literature and no protocols,” she says.“Everything has to be constructed from the ground up. Some days that feels hugely exciting – but other days it can feel like a mountain to climb.”
She was surprised to find that mammalian protocols worked well on plants.
“There are protocols for spatial transcriptomics in mammals. We weren’t expecting those to work, as mammalian protocols don’t always translate well into plants. But, the first time we tried one, we got results, which was a real surprise.”
In April this year the Institute was the first site in the UK to purchase the Vizgen MERSCOPE®.
The platform – based on multiplexed error-robust fluorescence in situ hybridization, or MERFISH – is the first commercially available platform to spatially profile and image the transcriptome at whole tissue section, single-cell, and sub-cellular levels.
Lister explains this allows researchers to look at cell organelle resolution without disturbing the positions of transcripts within the cell. This offers a unique insight into the activity of genes.
“Previous single cell work involved breaking the cell down into components and we sequenced them as individual units,” she says. “Here, a block of material is sliced into sections 10 microns thick and put on microscope slides.”
The sections therefore retain the position of the cell in the tissue, and the positions of everything in the cell. Activity is visualised using small DNA probes which bind to the products of 300 genes which could be active within the cell, linking phenotype and functionality.
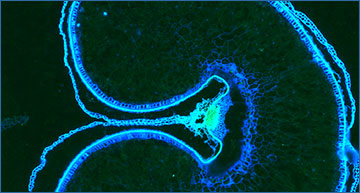
Wheat spikelet fluorescence using the Vizgen MERSCOPE® (©EarlhamInstitute/Ashleigh Lister)
The Earlham Institute boasts a dedicated laboratory supporting cutting-edge single-cell sequencing and analysis. It houses state-of-the-art platforms capable of genomic, epigenomic, and transcriptomic analyses of individual cells from a wide variety of organisms.
Spatial transcriptomics is just one of the tools the Institute has at its fingertips. Dr Iain Macaulay, Technical Development Group Leader at the Institute, stresses the importance of the accumulated expertise in the single-cell team as well as platforms and equipment.
“We are so fortunate to have a team of experienced and talented staff with extensive knowledge and expertise with these kinds of technologies,” he says.
“For example, Ashleigh has a background in both plant science and single cell analysis and she’s using that invaluable combination of experience to develop a completely new area of work for us.”
The innovative work being pioneered by Lister builds on the Institute’s National Bioscience Research Infrastructure (NBRI) in Transformative Genomics. It also feeds into part of the Institute’s Cellular Genomics research programme, designed to investigate the impact of cellular genomic and transcriptomic variation.
These technologies are key to unravelling how variation in sequence and regulation of the genome can contribute to an organism’s overall function and phenotype.
The Earlham Institute has the platforms and expertise to support all stages of a single- or multi-cell analysis – cell isolation, library preparation, sequencing and analysis – whether researchers are starting from tissue, sorted cells or pre-made libraries
“While we are still developing our skills in spatial transcriptomics, we are aiming to make it available to the wider community through collaboration or service provision,” Dr Macaulay adds.
“We would welcome enquiries from researchers or organisations looking to explore how this could benefit their own programmes of work – in plants, or indeed any other species.”
Source – Earlham Institute