The Drosophila eye has long been a valuable model for studying various biological processes such as differentiation, proliferation, apoptosis, and tissue morphogenesis. However, until now, there hasn’t been a comprehensive single-cell RNA sequencing (scRNA-seq) resource that captures the gene expression dynamics throughout the development of the larval eye disc. This gap in knowledge has made it challenging to understand the detailed processes that guide eye development at the cellular level.
In a groundbreaking study, researchers at the Baylor College of Medicine have now provided transcriptomic data from 13,000 individual cells, covering six key developmental stages of the larval eye. This data reveals the gene expression patterns from the initiation of the morphogenetic furrow—a crucial structure in eye development—to the differentiation of various photoreceptor cells and early cone cells. Let’s dive into what this means and why it’s important.
Understanding the Larval Eye Development Stages
The study tracks the development of the Drosophila eye at six distinct stages. This includes:
- Initiation of the Morphogenetic Furrow: This stage is where the process of eye development begins.
- Progression and Differentiation: As development continues, different types of photoreceptor cells and early cone cells start to form.
By analyzing gene expression at these stages, researchers can identify which genes are active at each point, providing insights into how cells transition from one type to another.
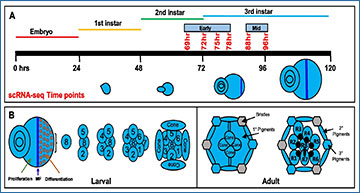
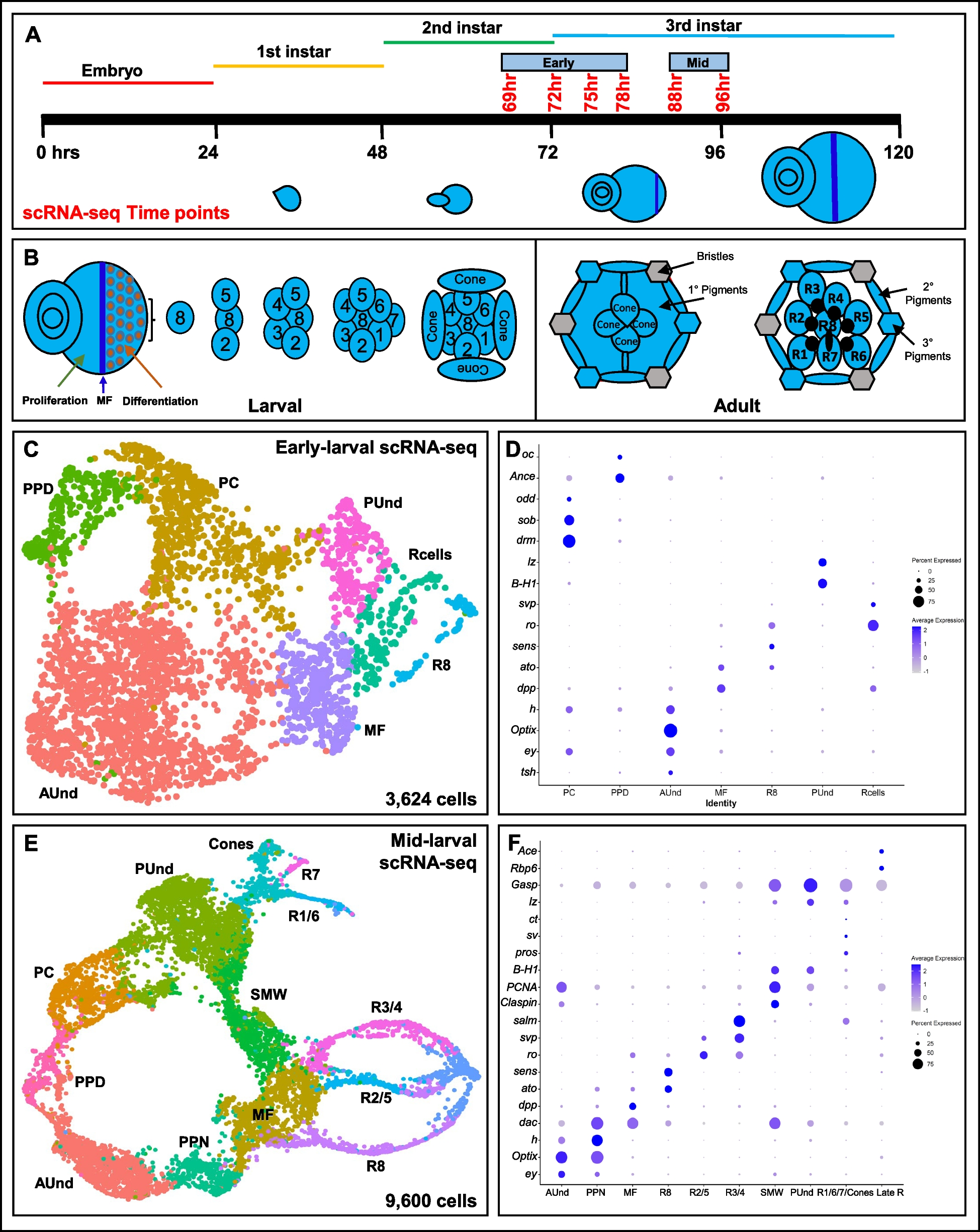
A Schematic of single cell RNA sequencing data generation from early to mid-larval eye discs. The time points reported in this work are shown in red. B Schematic of the larval eye disc depicting ommatidial assembly and different cell types present in the larval and adult retina. The morphogenetic furrow is shown as a blue vertical line across the eye disc separating the anterior proliferating cells and the differentiating and mature posterior cells. C scRNA-seq cluster plot generated from 3,624 early larval eye disc cells (69, 72, 75 and 78 h after egg laying (AEL)) shows all expected cell identities. The ‘Rcells’ cluster comprises the R2/5, R3/4 and R1/6 subtypes. D DotPlot showing the expression of known markers. The size of the dot indicates the percentage of cells in a cluster showing expression of a given gene, while the intensity of the blue color shows the average expression level across all cells within each cluster. E scRNA-seq cluster plot generated from 9,600 mid-larval (88 and 96 h AEL) eye disc cells with clusters corresponding to expected cell identities. F Dot plot showing the expression of mid-larval marker genes of different cell types. AUnd: Anterior Undifferentiated; PPN: Preproneural; MF: Morphogenetic Furrow; SMW: Second Mitotic Wave; PUnd: Posterior Undifferentiated; PC: Posterior Cuboidal Margin Peripodium; and PPD: Anterior Peripodial
Key Findings: Cell Clusters and Gene Expression
The analysis identified clusters of cells that correspond to all major cell types in the eye disc. This means that researchers can now see how different cells in the eye disc evolve over time. More importantly, the study uncovered dozens of genes specific to certain cell types that had not been previously reported to play a role in eye development.
These discoveries are significant because they:
- Highlight New Genes: Identifying new genes involved in eye development opens up new avenues for research. Scientists can now explore these genes to understand their specific roles and how they contribute to the formation and function of the eye.
- Enhance Understanding of Cell Differentiation: By understanding which genes are active at different stages, researchers can better understand how undifferentiated cells become specialized cells, such as photoreceptors and cone cells.
Implications for Future Research
This single-cell RNA sequencing data is a treasure trove for researchers. It provides a detailed map of gene expression throughout the critical stages of eye development in Drosophila. This resource will be invaluable for studying various aspects of eye development, such as:
- Gene Function: Researchers can investigate the newly identified genes to determine their specific functions in eye development.
- Developmental Biology: The data helps in understanding the general principles of how tissues and organs develop at the cellular level.
- Model System Insights: The Drosophila eye serves as a model system for other organisms, including humans. Insights gained from this study could have broader implications for understanding eye development and diseases in other species.
The release of this single-cell RNA sequencing data marks a significant advancement in our understanding of eye development in Drosophila. By capturing gene expression dynamics across six developmental stages, this study provides a detailed and comprehensive view of how the eye forms and differentiates at the cellular level. Researchers now have access to a powerful resource that will facilitate deeper insights into the mechanisms of eye development, potentially leading to breakthroughs in both basic biology and medical research.
This new data not only enriches our understanding of the Drosophila eye but also underscores the value of single-cell RNA sequencing in uncovering the complexities of developmental biology.
Raja KKB, Yeung K, Li Y, Chen R, Mardon G. (2024) A single cell RNA sequence atlas of the early Drosophila larval eye. BMC Genomics 25(1):616. [article]