Clinical manifestation of prostate cancer (PCa) is highly variable. Aggressive tumors require radical treatment while clinically non-significant ones may be suitable for active surveillance. Researchers at the Fraunhofer Institute for Cell Therapy and Immunology previously developed the prognostic ProstaTrend RNA signature based on transcriptome-wide microarray and RNA-sequencing (RNA-Seq) analyses, primarily of prostatectomy specimens. An RNA-Seq study of formalin-fixed paraffin-embedded (FFPE) tumor biopsies has now allowed the researchers to use this test as a basis for the development of a novel test that is applicable to FFPE biopsies as a tool for early routine PCa diagnostics.
All patients of the FFPE biopsy cohort were treated by radical prostatectomy and median follow-up for biochemical recurrence (BCR) was 9 years. Based on the transcriptome data of 176 FFPE biopsies, the researchers filtered ProstaTrend for genes susceptible to FFPE-associated degradation via regression analysis. ProstaTrend was additionally restricted to genes with concordant prognostic effects in the RNA-Seq TCGA prostate adenocarcinoma (PRAD) cohort to ensure robust and broad applicability. The prognostic relevance of the refined Transcriptomic Risk Score (TRS) was analyzed by Kaplan-Meier curves and Cox-regression models in our FFPE-biopsy cohort and 9 other public datasets from PCa patients with BCR as primary endpoint. In addition, they developed a prostate single-cell atlas of 41 PCa patients from 5 publicly available studies to analyze gene expression of ProstaTrend genes in different cell compartments.
Validation of the TRS using the original ProstaTrend signature in the cohort of FFPE biopsies revealed a relevant impact of FFPE-associated degradation on gene expression and consequently no significant association with prognosis (Cox-regression, p-value > 0.05) in FFPE tissue. However, the TRS based on the new version of the ProstaTrend-ffpe signature, which included 204 genes (of originally 1396 genes), was significantly associated with BCR in the FFPE biopsy cohort (Cox-regression p-value < 0.001) and retained prognostic relevance when adjusted for Gleason Grade Groups. The researchers confirmed a significant association with BCR in 9 independent cohorts including 1109 patients. Comparison of the prognostic performance of the TRS with 17 other prognostically relevant PCa panels revealed that ProstaTrend-ffpe was among the best-ranked panels. They generated a PCa cell atlas to associate ProstaTrend genes with cell lineages or cell types. Tumor-specific luminal cells have a significantly higher TRS than normal luminal cells in all analyzed datasets. In addition, TRS of epithelial and luminal cells was correlated with increased Gleason score in 3 studies.
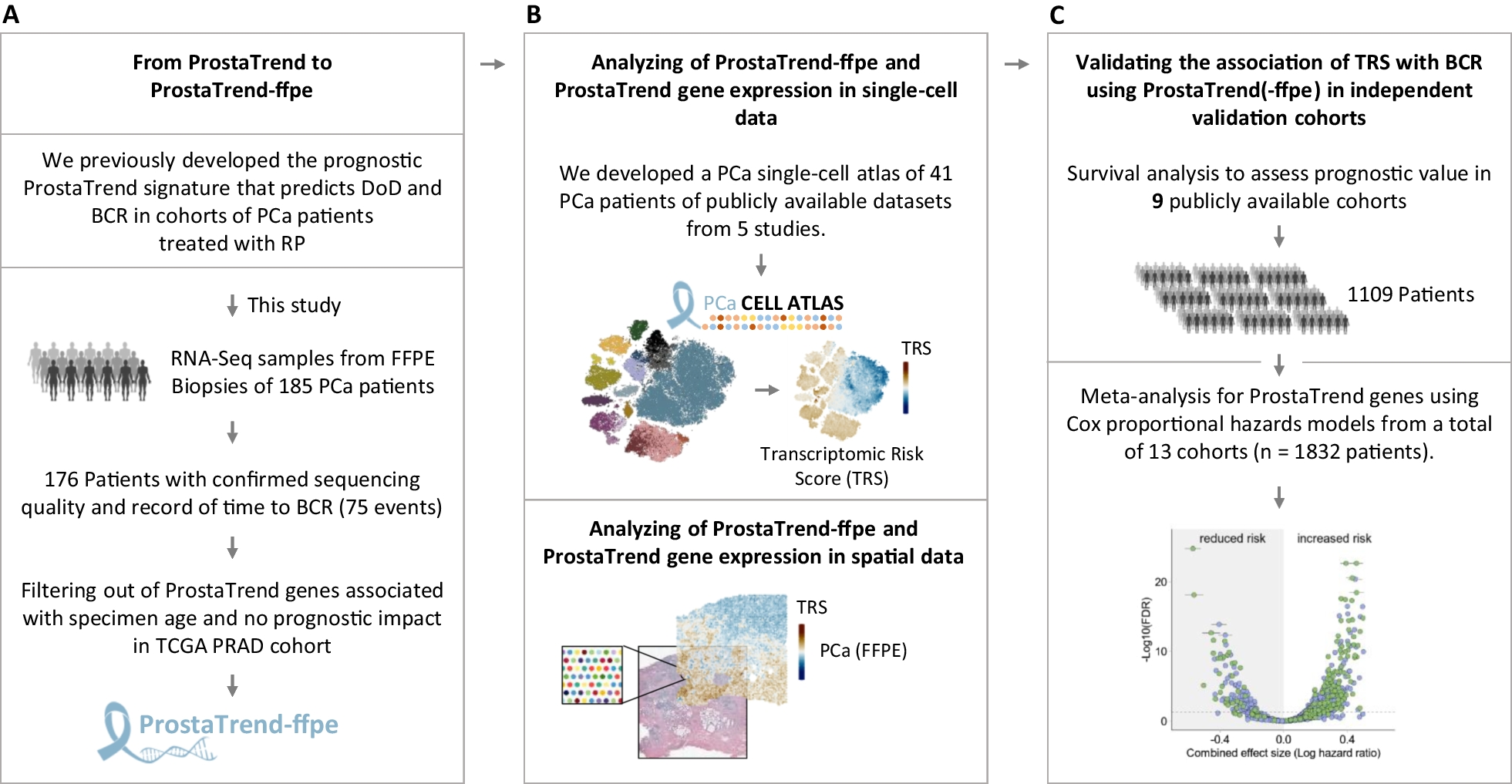
Analysis workflow
A Overview of the included tissue samples we used to develop the ProstaTrend-ffpe signature. Shown are the number of patients included in the study and reasons for exclusion. For 185 patients, we performed a strand-specific transcriptome-wide sequencing from FFPE biopsy tissue. A total of six samples did not meet the quality criteria for RNA-Seq data. In addition, we excluded three samples due to missing clinical follow-up data. The final cohort included 176 patients, for 75 of whom BCR was observed within the follow-up time. FFPE formalin-fixed paraffin-embedded, PCa prostate cancer, DoD death of disease, BCR biochemical recurrence. B For the development of the PCa single-cell atlas, we used scRNA-Seq data of PCa patients from 5 publicly available studies. Spatial transcriptomics data of a human PCa biopsy (GS = 3 + 4) were downloaded from the 10× Genomics database. C The prognostic value of the Transcriptomic Risk Scores (TRS) using ProstaTrend(-ffpe) was evaluated by survival analyses in 9 publicly available cohorts and a meta-analysis with a total of 13 cohorts
Availability – An interactive version of the results of this study can be accessed at https://bioinf.izi.fraunhofer.de/prostatrend/.
Rade M, Kreuz M, Borkowetz A, Sommer U, Blumert C, Füssel S, Bertram C, Löffler D, Otto DJ, Wöller LA, Schimmelpfennig C, Köhl U, Gottschling AC, Hönscheid P, Baretton GB, Wirth M, Thomas C, Horn F, Reiche K. (2024) A reliable transcriptomic risk-score applicable to formalin-fixed paraffin-embedded biopsies improves outcome prediction in localized prostate cancer. Mol Med. 30(1):19. [article]