RNA editing is a post-transcriptional modification with a cell-specific manner and important biological implications. Although single-cell RNA-seq (scRNA-seq) is an effective method for studying cellular heterogeneity, it is difficult to detect and study RNA editing events from scRNA-seq data because of the low sequencing coverage. To overcome this, researchers at the University of Chinese Academy of Sciences have developed a computational method to systematically identify RNA editing sites of cell types from scRNA-seq data. To demonstrate its effectiveness, the researchers apply it to scRNA-seq data of human hematopoietic stem/progenitor cells (HSPCs) with an annotated lineage differentiation relationship according to previous research and study the impacts of RNA editing on hematopoiesis. The dynamic editing patterns reveal the relevance of RNA editing on different HSPCs. For example, four microRNA (miRNA) target sites on 3ʹ UTR of EIF2AK2 are edited across all HSPC populations, which may abolish the miRNA-mediated inhibition of EIF2AK2. Elevated EIF2AK2 may thus activate the integrated stress response (ISR) pathway to initiate global translational attenuation as a protective mechanism to maintain cellular homeostasis during HSPCs’ differentiation. These findings also indicate that RNA editing plays an essential role in the coordination of lineage commitment and self-renewal of hematopoietic stem cells (HSCs). Taken together, these researchers demonstrate the capacity of scRNA-seq data to exploit RNA editing events of cell types, and find that RNA editing may exert multiple modules of regulation in hematopoietic processes.
A schematic diagram showing the computational method developed in this study
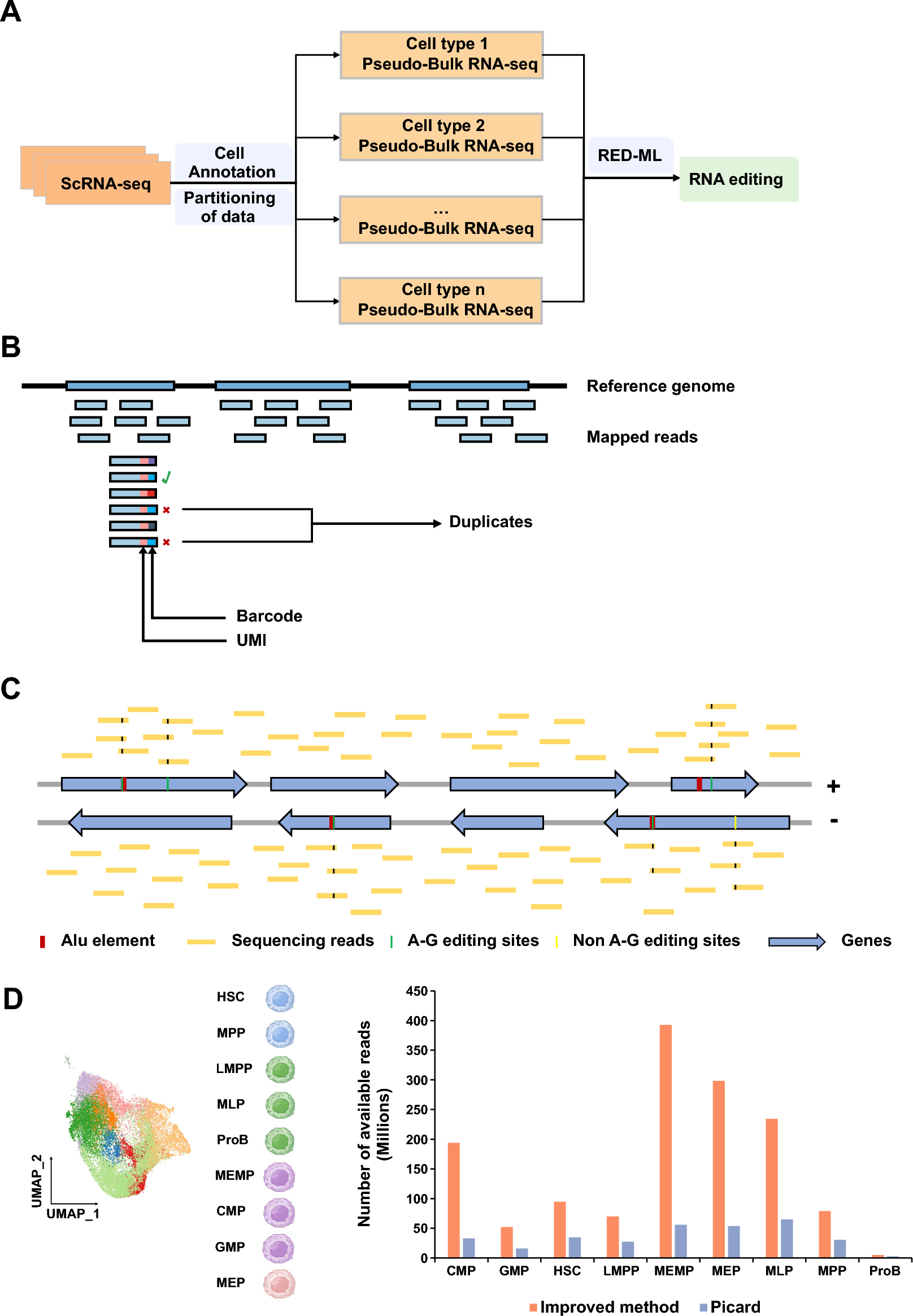
(A) A workflow showing the strategies to identify RNA editing events using scRNA-seq. The cell type annotation information was used to combine the mapped reads of the same cell type in scRNA-seq to obtain pseudo-Bulk RNA-seq for each cell type. (B) The novel threshold used to mark duplicates. Only if the aligned reads with the same alignment position, UMI and barcode are defined as duplicates. (C) The reads aligned to the reference genome are divided into reads aligned to the forward strand and reverse strand to distinguish RNA editing sites occurring on the forward/reverse strand genes. The edited sites located in ALU element and with the A-G variation are more likely to be identified as RNA editing sites. (D) The scRNA-seq data of hematopoietic stem cells (HSCs), multipotential progenitor cells (MPPs), lymphoid lineage multipotential progenitor cells (LMPPs), multipotential lymphoid lineage progenitor cells (MLPs), megakaryocyte erythroid mast progenitor cells (MEMPs), common myeloid progenitor cells (CMPs), granulocyte monocyte progenitors (GMPs), megakaryocyte erythroid progenitors (MEPs), and B-cell progenitors (ProBs) were used to evaluate improved method and Picard. Bar plot showing the number of available reads using improved method and Picard.
Wu Y, Hao S, Xu X, Dong G, Ouyang W, Liu C, Sun HX. (2023) A novel computational method enables RNA editome profiling during human hematopoiesis from scRNA-seq data. Sci Rep 13(1):10335. [article]