RNA sequencing (RNA-Seq) is a cutting-edge technique that’s gaining traction in clinical research and drug development. It’s a method used to study the quantity and sequences of RNA in a sample, providing insights into gene expression and helping to uncover molecular mechanisms of diseases. However, different RNA-Seq methods have unique advantages, especially when dealing with challenging samples like degraded RNA or low-input RNA (RNA collected in small amounts, often in clinical settings).
Standard RNA-Seq Method: A Quick Overview
The traditional approach to RNA-Seq involves capturing mRNA (messenger RNA) using poly(A) tails, a sequence of adenine nucleotides at the end of mRNA molecules. This method uses Oligo dT beads to bind these tails and isolate the mRNA. While effective for high-quality RNA, this technique isn’t suitable for degraded RNA, which often lacks intact poly(A) tails.
Alternative RNA-Seq Methods
To address the limitations of the standard method, researchers have developed alternative RNA-Seq library preparation kits that use random primers instead of Oligo dT beads. This study focuses on three such methods:
- SMART-Seq
- xGen Broad-range
- RamDA-Seq
These methods aim to capture RNA more effectively, even when it’s degraded or present in low amounts.
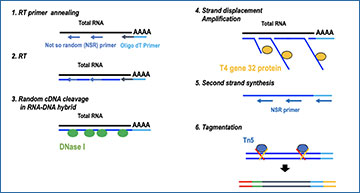
Library preparation workflow
(A) SMART-Seq, (B) xGen, and (C) RamDA-Seq
Evaluating Performance
Researchers at Kanazawa Medical University Hospital compared these alternative methods to the standard RNA-Seq technique by examining several factors:
- Correlation: How closely the results match the expected gene expression patterns.
- Number of Detected Expressing Genes: The total number of genes that show activity.
- Expression Levels: The intensity of gene activity signals.
Here’s what they found:
- RamDA-Seq: This method performed similarly to the standard RNA-Seq in terms of correlation and detected gene numbers. However, its performance dropped with low-input and degraded RNA.
- xGen Broad-range: This method didn’t perform as well as SMART-Seq, especially with challenging RNA samples.
- SMART-Seq: This method stood out by showing better performance with low-input and degraded RNA samples compared to both xGen and RamDA-Seq.
Ribosomal RNA (rRNA) Depletion
Ribosomal RNA, which makes up a large portion of total RNA, can interfere with the detection of mRNA. By depleting rRNA from samples, the researchers found that the performance of both SMART-Seq and xGen improved, as this increased the expression levels of other RNA types.
Conclusion: SMART-Seq with rRNA Depletion
For RNA-Seq applications involving low-input or degraded RNA, the SMART-Seq method, especially when combined with rRNA depletion, offers significant advantages. This makes it a valuable tool in clinical settings where sample quality and quantity are often less than ideal.
In summary, understanding the strengths and limitations of different RNA-Seq methods allows researchers to choose the best approach for their specific needs. SMART-Seq, particularly with rRNA depletion, emerges as a robust choice for handling challenging RNA samples, paving the way for more accurate and insightful gene expression analysis in clinical research and drug development.
Ura H, Niida Y. Comparison of RNA-Sequencing Methods for Degraded RNA. Int J Mol Sci 25(11):6143. [article]