Tissues are highly complicated with spatial heterogeneity in gene expression. However, the cutting-edge single-cell RNA-seq technology eliminates the spatial information of individual cells, which contributes to the characterization of cell identities. Zhejiang University researchers have developed a single-cell spatial position associated co-embeddings (scSpace), an integrative method to identify spatially variable cell subpopulations by reconstructing cells onto a pseudo-space with spatial transcriptome references (Visium, STARmap, Slide-seq, etc.). The researchers benchmarked scSpace with both simulated and biological datasets, and demonstrate that scSpace can accurately and robustly identify spatially variated cell subpopulations. When employed to reconstruct the spatial architectures of complex tissue such as the brain cortex, the small intestinal villus, the liver lobule, the kidney, the embryonic heart, and others, scSpace shows promising performance on revealing the pairwise cellular spatial association within single-cell data. The application of scSpace in melanoma and COVID-19 exhibits a broad prospect in the discovery of spatial therapeutic markers.
Schematic workflow of scSpace and performance evaluations on simulated data
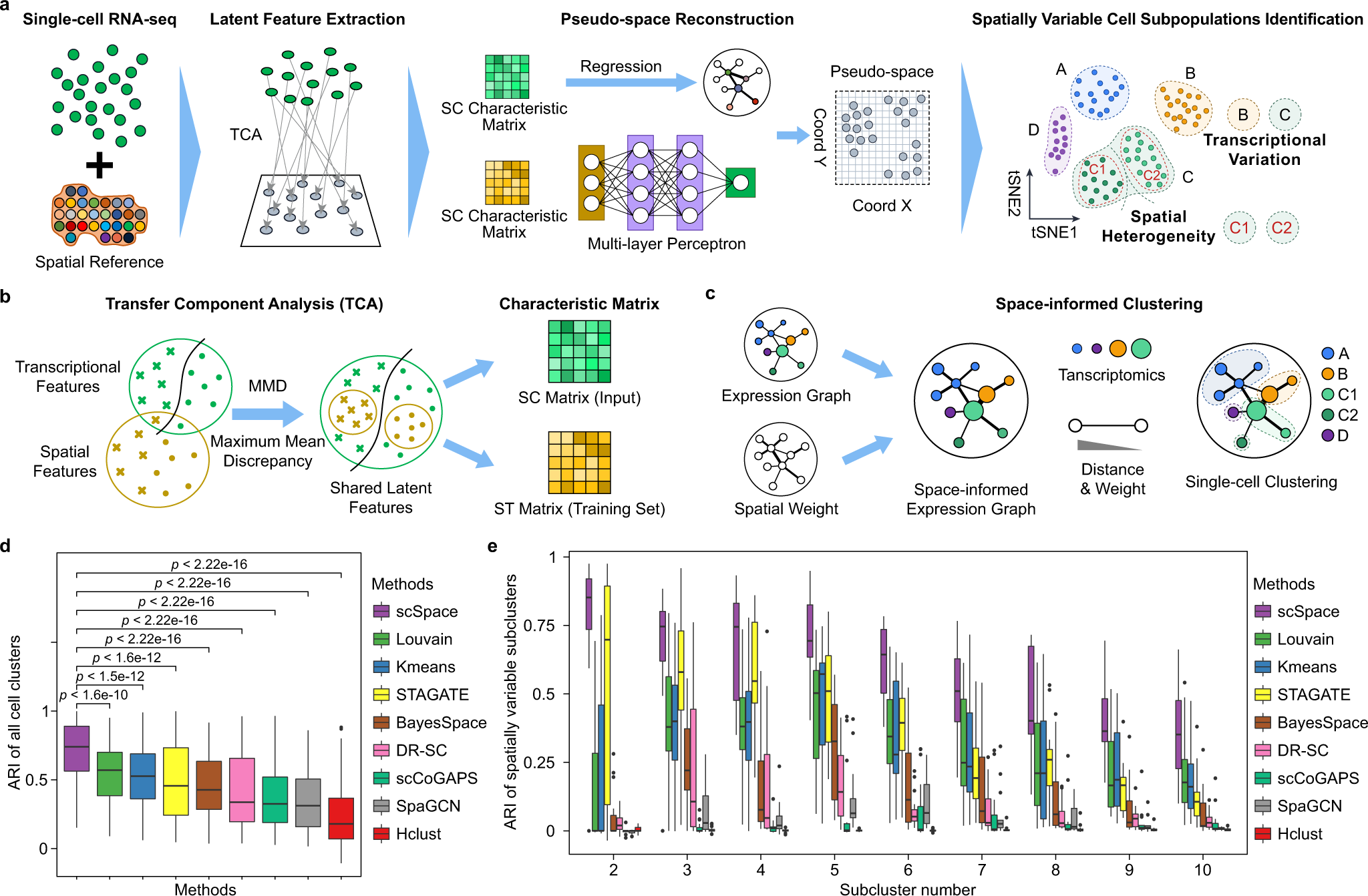
a Overview of the design concept of scSpace. Given the scRNA-seq data (SC) and spatial transcriptomics reference (ST), scSpace co-embeds these two types of data into a shared latent space and extracts the shared latent features. Using the characteristic matrix from ST data, scSpace trains a multi-layer perceptron model with spatial coordinates as the outcome and latent features as the predictors. The trained model is then applied to the characteristic matrix from SC data for pseudo-space reconstruction. Based on the gene expression profiles as well as the pseudo-space information, scSpace identifies the spatially variable cell subpopulations from scRNA-seq data. b Conceptual framework of latent feature extraction with scSpace. A transfer learning method termed transfer component analysis (TCA) is applied to extract the shared latent feature representation across scRNA-seq and spatial transcriptomics data. TCA first projects the scRNA-seq and spatial transcriptomics data into a Reproducing Kernel Hilbert Space (RKHS), and then reduce the difference in the distribution of transformed two domain data by minimizing the maximum mean discrepancy (MMD) between them. The shared latent feature representation across two domain data is then extracted for the next pseudo-space reconstruction step. c Conceptual framework of space-informed clustering with scSpace. The gene expression graph is first constructed on the reduced principal components derived from normalized gene expression profiles of single cells using the k-nearest neighbor (KNN) algorithm. For each edge in the gene expression graph, a spatial weight is introduced based on the distances between cells in the pseudo-space. Then, scSpace performs the unsupervised clustering step on the space-informed gene expression graph to identify spatially variable cell subpopulations from scRNA-seq data. d and e Comparison of scSpace with other existing clustering methods in identifying all cell clusters (d) and only spatially heterogeneous subclusters (e) on 140 simulated datasets. Data are presented as boxplots (minima, 25th percentile, median, 75th percentile, and maxima). P-value is calculated with the two-sided Wilcoxon rank-sum test (the exact P-values from left to right are 1.6e−10, 1.5e−12, 1.6e−12, 3.1e−18, 1.1e−17, 4.7e−28, 2.2e−30, and 4.0e−35, respectively).
Availability – The scSpace algorithm and related analysis are available at: https://github.com/ZJUFanLab/scSpace.
Qian J, Liao J, Liu Z, Chi Y, Fang Y, Zheng Y, Shao X, Liu B, Cui Y, Guo W, Hu Y, Bao H, Yang P, Chen Q, Li M, Zhang B, Fan X. (2023) Reconstruction of the cell pseudo-space from single-cell RNA sequencing data with scSpace. Nat Commun 14(1):2484. [article]