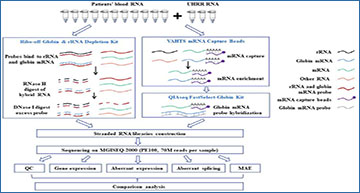
As an adjunct to diagnostic exome sequencing and whole-genome sequencing, RNA sequencing (RNA-seq) has been demonstrated to improve diagnostic yield for Mendelian diseases. However, systematic evaluation of the associated experimental and computational processes and the establishment of robust and efficient practices for RNA diagnostics implemented in the clinic to analyse readily accessible whole blood samples are still required.
Researchers at the University of Chinese Academy of Sciences simulated clinical conditions in which each patient’s sample is tested only once, and they evaluated the two typical experimental protocols (polyA-selection and rRNA depletion) by comparing the expression profiles, aberrant splicing events and monoallelic expression (MAE) identified from 11 patients in clinical settings with different bioinformatics software.
Study design
Blood RNA from 11 patients with suspected hereditary neuromuscular disorders and 3 replicates of Universal Human Reference RNA (UHRR) were performed in parallel by polyA-selection (polyA+) and rRNA depletion (ribo-minus). The expression profiles, aberrant splicing and monoallelically expressed (MAE) rare variants derived from the two methods were compared to determine which method is most suitable for genetic diagnosis and to establish standardized practices for RNA diagnostics using clinically accessible whole blood samples.
The researchers demonstrated that a higher proportion of unique reads from polyA-selection than rRNA depletion were mapped to exons or exon – intron junction regions (84.54% vs. 40.14%), resulting in more detectable OMIM genes (TPM > 1) in the blood (65.29% vs. 59.79%); thus, the rRNA depletion method requires a median of 258 more valid reads per gene to achieve the same level of gene quantification. Moreover, although the transcriptome profiling of protein-coding genes in the two methods is highly correlated, polyA-selection offers more sensitive detection of MAE variants and aberrant splicing under common filtering conditions in combination with DROP.
A combination of polyA+ and DROP is recommended when implementing blood-based RNA-seq for the diagnosis of Mendelian diseases in clinical practice, and filtering criteria for aberrant expression, aberrant splicing and MAE variants are suggested for reference.
Yang Z, Yang X, Chen Y, Wang Z, Song L, Sun J, Peng H, Yang X, Peng Z, Yi Dai Y. (2023) Systematic evaluation of the two main blood-based RNA-seq approaches for Mendelian disease diagnosis. medRXiv [online pre-print]. [article]