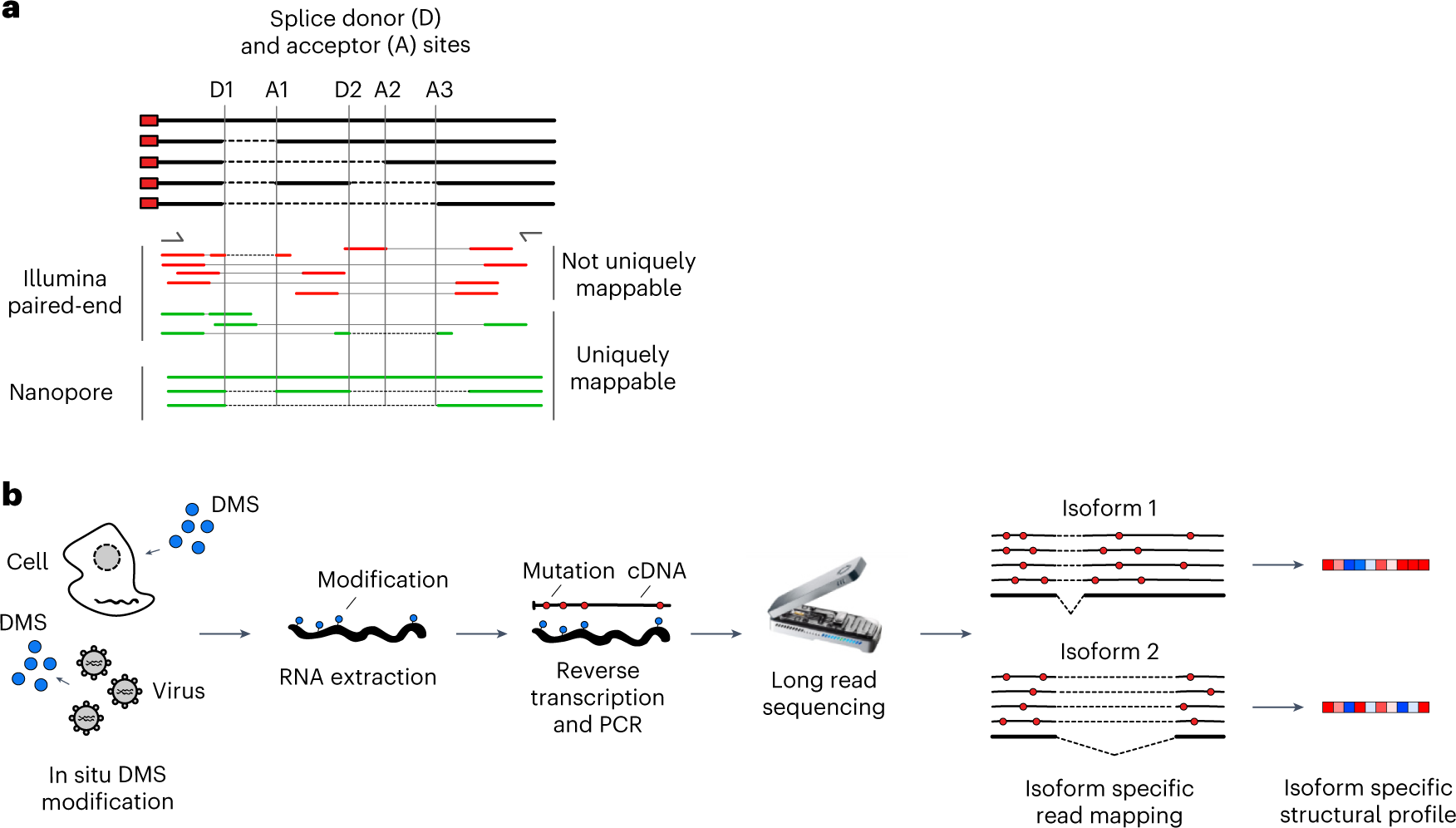
Genome-wide measurements of RNA structure can be obtained using reagents that react with unpaired bases, leading to adducts that can be identified by mutational profiling on next-generation sequencing machines. One drawback of these experiments is that short sequencing reads can rarely be mapped to specific transcript isoforms. Consequently, information is acquired as a population average in regions that are shared between transcripts, thus blurring the underlying structural landscape.
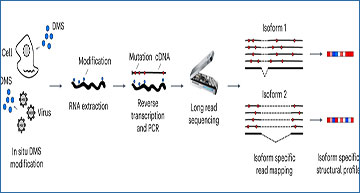
Researchers at the Helmholtz Institute for RNA-based Infection Research have developed nanopore dimethylsulfate mutational profiling (Nano-DMS-MaP)—a method that exploits long-read sequencing to provide isoform-resolved structural information of highly similar RNA molecules. The researchers demonstrate the value of Nano-DMS-MaP by resolving the complex structural landscape of human immunodeficiency virus-1 transcripts in infected cells. They show that unspliced and spliced transcripts have distinct structures at the packaging site within the common 5′ untranslated region, likely explaining why spliced viral RNAs are excluded from viral particles. Thus, Nano-DMS-MaP is a straightforward method to resolve biologically important transcript-specific RNA structures that were previously hidden in short-read ensemble analyses.
Transcript isoforms structural analysis by Nano-DMS-MaP
Nano-DMS-MaP for isoform-resolved RNA structure determination. a, Transcript isoforms can be generated by alternative splicing. Common regions between transcript isoforms may have different structures, but short sequencing reads cannot always be unambiguously mapped to transcript isoforms. Long-read nanopore sequencing can be uniquely mapped to transcript isoforms. b, DMS can probe RNA structure in situ in cells or virions. Modified RNA is extracted and reverse transcribed into long cDNA molecules for sequencing on nanopore devices. Isoform-specific read mapping allows isoform-specific structural profiling. MinION device image credit: Oxford Nanopore Technologies (2023).
Availability – Code used for the Nano-DMS-MaP analysis is accessible via the Smyth-lab Github (https://github.com/smyth-lab/Nano-DMS-MaP).
Bohn P, Gribling-Burrer AS, Ambi UB, Smyth RP. (2023) Nano-DMS-MaP allows isoform-specific RNA structure determination. Nat Methods [Epub ahead of print]. [article]