Characterizing the topology of gene regulatory networks (GRNs) is a fundamental problem in systems biology. The advent of single cell technologies has made it possible to construct GRNs at finer resolutions than bulk and microarray datasets. However, cellular heterogeneity and sparsity of the single cell datasets render void the application of regular Gaussian assumptions for constructing GRNs. Additionally, most GRN reconstruction approaches estimate a single network for the entire data. This could cause potential loss of information when single cell datasets are generated from multiple treatment conditions/disease states.
To better characterize single cell GRNs under different but related conditions, Michigan State University researchers propose the joint estimation of multiple networks using multiple signed graph learning (scMSGL). The proposed method is based on recently developed graph signal processing (GSP) based graph learning, where GRNs and gene expressions are modeled as signed graphs and graph signals, respectively. scMSGL learns multiple GRNs by optimizing the total variation of gene expressions with respect to GRNs while ensuring that the learned GRNs are similar to each other through regularization with respect to a learned signed consensus graph. The researchers further kernelize scMSGL with the kernel selected to suit the structure of single cell data.
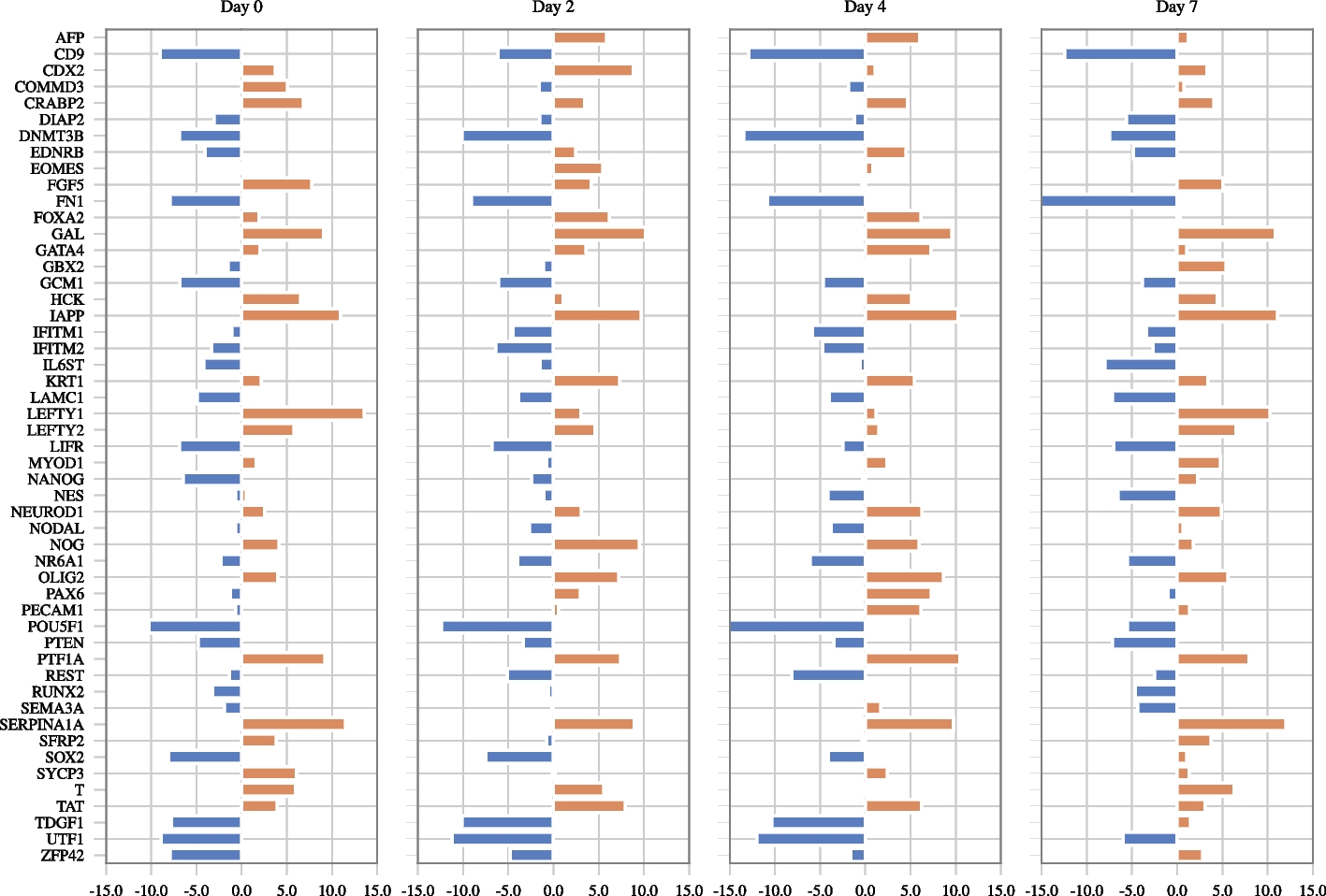
Genes with the highest node degrees
Orange and blue bars indicate that the degree is calculated using activating and inhibitory edges, respectively. Only genes whose activating or inhibitory degrees is among the top 15 genes in any view are shown
scMSGL is shown to have superior performance over existing state of the art methods in GRN recovery on simulated datasets. Furthermore, scMSGL successfully identifies well-established regulators in a mouse embryonic stem cell differentiation study and a cancer clinical study of medulloblastoma.
Availability – The scMSGL code and simulated data are available at https://github.com/Single-Cell-Graph-Learning/scMSGL.
Karaaslanli A, Saha S, Maiti T, Aviyente S. (2023) Kernelized multiview signed graph learning for single-cell RNA sequencing data. BMC Bioinformatics 24(1):127. [article]