RNA velocity provides an approach for inferring cellular state transitions from single-cell RNA sequencing (scRNA-seq) data. Conventional RNA velocity models infer universal kinetics from all cells in an scRNA-seq experiment, resulting in unpredictable performance in experiments with multi-stage and/or multi-lineage transition of cell states where the assumption of the same kinetic rates for all cells no longer holds.
Researchers at the Houston Methodist Research Institute have developed cellDancer, a scalable deep neural network that locally infers velocity for each cell from its neighbors and then relays a series of local velocities to provide single-cell resolution inference of velocity kinetics. In the simulation benchmark, cellDancer shows robust performance in multiple kinetic regimes, high dropout ratio datasets and sparse datasets. The researchers show that cellDancer overcomes the limitations of existing RNA velocity models in modeling erythroid maturation and hippocampus development. Moreover, cellDancer provides cell-specific predictions of transcription, splicing and degradation rates, which we identify as potential indicators of cell fate in the mouse pancreas.
Predicting RNA velocity in localized cell populations via DNNs
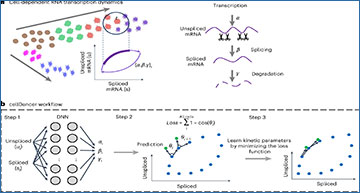
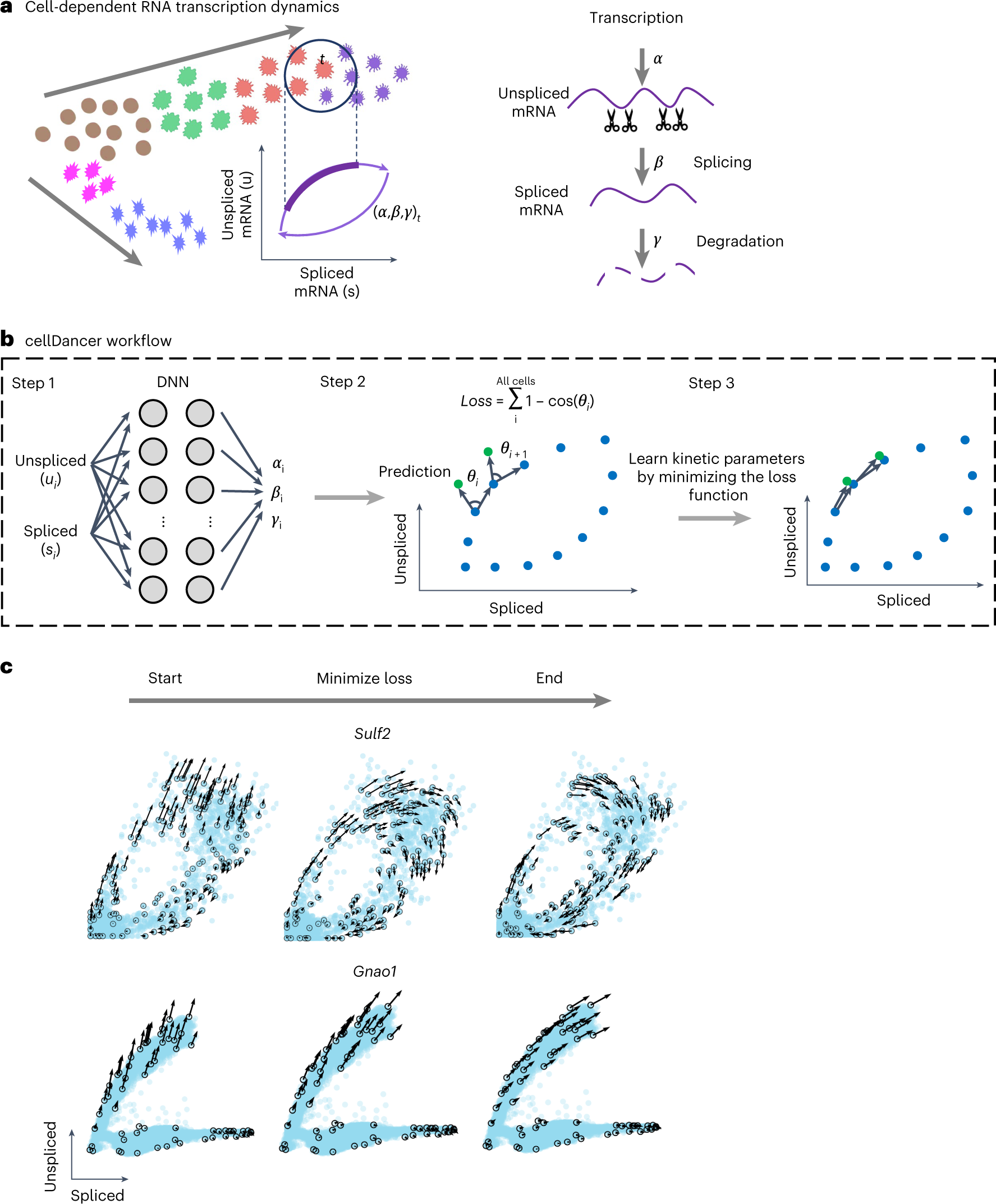
a, Transcription dynamics of the premature (unspliced) and mature (spliced) mRNAs are governed by the transcription (α), splicing (β) and degradation (γ) rates. Multi-kinetics genes involve multiple-lineage and/or multi-stage transitions of the cellular states; hence, cell-dependent rates (α, β, γ)t are required to accurately capture the transcription dynamics of those genes. In the illustration, the (α, β, γ)t for cell t are computed by locating the future state cell in the neighboring cells of t (‘local environment’), assuming that the cells in the local environment share the same (α, β, γ). b, cellDancer uses a DNN to predict cell-specific α, β and γ for each gene. The DNN consists of an input layer with the spliced and unspliced mRNA abundances (ui, si) i = 1,2, …, ncells, two fully connected hidden layers each with 100 nodes and an output layer yielding cell-specific α, β and γ. The loss function is defined as the sum of every cell’s cosine similarity of predicted and observed velocity vectors. The DNN is iteratively optimized by minimizing the loss function. c, The progress of minimizing the loss function. RNA velocities for the examples of the mono-kinetic gene Sulf2 in pancreatic endocrinogenesis, and the multi-lineage gene Gnao1 in mouse hippocampus maturation is projected onto the phase portraits during the training process of their DNNs.
Availability – cellDancer is implemented in Python and is available at https://github.com/GuangyuWangLab2021/cellDancer.
Li S, Zhang P, Chen W, Ye L, Brannan KW, Le NT, Abe JI, Cooke JP, Wang G. (2023) A relay velocity model infers cell-dependent RNA velocity. Nat Biotechnol [Epub ahead of print]. [article]