With the emergence of hundreds of single-cell RNA-sequencing (scRNA-seq) datasets, the number of computational tools to analyze aspects of the generated data has grown rapidly. As a result, there is a recurring need to demonstrate whether newly developed methods are truly performant-on their own as well as in comparison to existing tools. Benchmark studies aim to consolidate the space of available methods for a given task and often use simulated data that provide a ground truth for evaluations, thus demanding a high quality standard results credible and transferable to real data.
Researchers at the University of Zurich evaluated methods for synthetic scRNA-seq data generation in their ability to mimic experimental data. Besides comparing gene- and cell-level quality control summaries in both one- and two-dimensional settings, the researchers further quantified these at the batch- and cluster-level. Secondly, they investigate the effect of simulators on clustering and batch correction method comparisons, and, thirdly, which and to what extent quality control summaries can capture reference-simulation similarity.
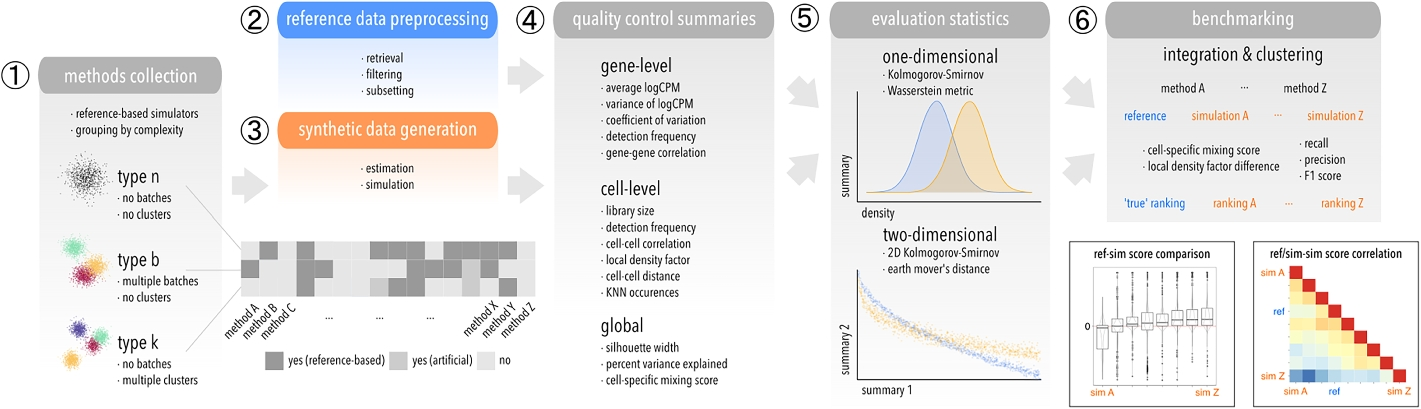
Schematic of the computational workflow used to benchmark scRNA-seq simulators
(1) Methods are grouped according to which level of complexity they can accommodate: type n (“singular”), b (batches), k (clusters). (2) Raw datasets are retrieved reproducibly from a public source, filtered, and subsetted into various datasets that serve as reference for (3) parameter estimation and simulation. (4) Various gene-, cell-level, and global summaries are computed from reference and simulated data, and (5) compared in a one- and two-dimensional setting using two statistics each. (6) Integration and clustering methods are applied to type b and k references and simulations, respectively, and relative performances compared between reference-simulation and simulation-simulation pairs
These results suggest that most simulators are unable to accommodate complex designs without introducing artificial effects, they yield over-optimistic performance of integration and potentially unreliable ranking of clustering methods, and it is generally unknown which summaries are important to ensure effective simulation-based method comparisons.
Crowell HL, Morillo Leonardo SX, Soneson C, Robinson MD. (2023) The shaky foundations of simulating single-cell RNA sequencing data. Genome Biol 24(1):62. [article]