The recent development of single-cell techniques is essential to unravel complex biological systems. By measuring the transcriptome and the accessible genome on a single-cell level, cellular heterogeneity in a biological environment can be deciphered. Transcription factors act as key regulators activating and repressing downstream target genes, and together they constitute gene regulatory networks that govern cell morphology and identity. Dissecting these gene regulatory networks is crucial for understanding molecular mechanisms and disease, especially within highly complex biological systems. The gene regulatory network analysis software ANANSE and the motif enrichment software GimmeMotifs were both developed to analyse bulk datasets.
Radboud University researchers have developed scANANSE, a software pipeline for gene regulatory network analysis and motif enrichment using single-cell RNA and ATAC datasets. The scANANSE pipeline can be run from either R or Python. First, it exports data from standard single-cell objects. Next, it automatically runs multiple comparisons of cell cluster data. Finally, it imports the results back to the single-cell object, where the result can be further visualised, integrated, and interpreted. Here, the researchers demonstrate their scANANSE pipeline on a publicly available PBMC multi-omics dataset. It identifies well-known cell type-specific hematopoietic factors. Importantly, they also demonstrated that scANANSE combined with GimmeMotifs is able to predict transcription factors with both activating and repressing roles in gene regulation.
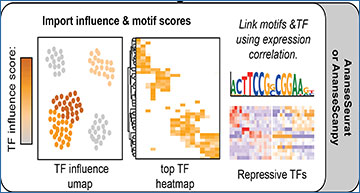
An overview of the single-cell ANANSE pipeline
After pre-processing and clustering, data is exported using either AnanseSeurat or AnanseScanpy. Next, Anansnake automatically runs ANANSE after which the influence scores and motif enrichment results with AnanseSeurat or AnanseScanpy are imported. In parallel, Anansnake runs motif enrichment analysis using gimme maelstrom, and the motif results are imported and linked to the highest correlating TFs using the single-cell object scRNA-seq data.
Smits JGA, Arts JA, Frölich S, Snabel RR, Heuts BMH, Martens JHA, van HeeringenSJ, Zhou H. (2023) scANANSE gene regulatory network and motif analysis of single-cell clusters. F1000Research [Epub ahead of print]. [article]