Natural sequence variation within mitochondrial DNA (mtDNA) contributes to human phenotypes and may serve as natural genetic markers in human cells for clonal and lineage tracing. Stanford University researchers have developed a single-cell multi-omic approach, called ‘mitochondrial single-cell assay for transposase-accessible chromatin with sequencing’ (mtscATAC-seq), enabling concomitant high-throughput mtDNA genotyping and accessible chromatin profiling. Specifically, this technique allows the mitochondrial genome-wide inference of mtDNA variant heteroplasmy along with information on cell state and accessible chromatin variation in individual cells. Leveraging somatic mtDNA mutations, this method further enables inference of clonal relationships among native ex vivo-derived human cells not amenable to genetic engineering-based clonal tracing approaches.
Here, the researchers provide a step-by-step protocol for the use of mtscATAC-seq, including various cell-processing and flow cytometry workflows, by using primary hematopoietic cells, subsequent single-cell genomic library preparation and sequencing that collectively take ~3–4 days to complete. They discuss experimental and computational data quality control metrics and considerations for the extension to other mammalian tissues. Overall, mtscATAC-seq provides a broadly applicable platform to map clonal relationships between cells in human tissues, investigate fundamental aspects of mitochondrial genetics and enable additional modes of multi-omic discovery.
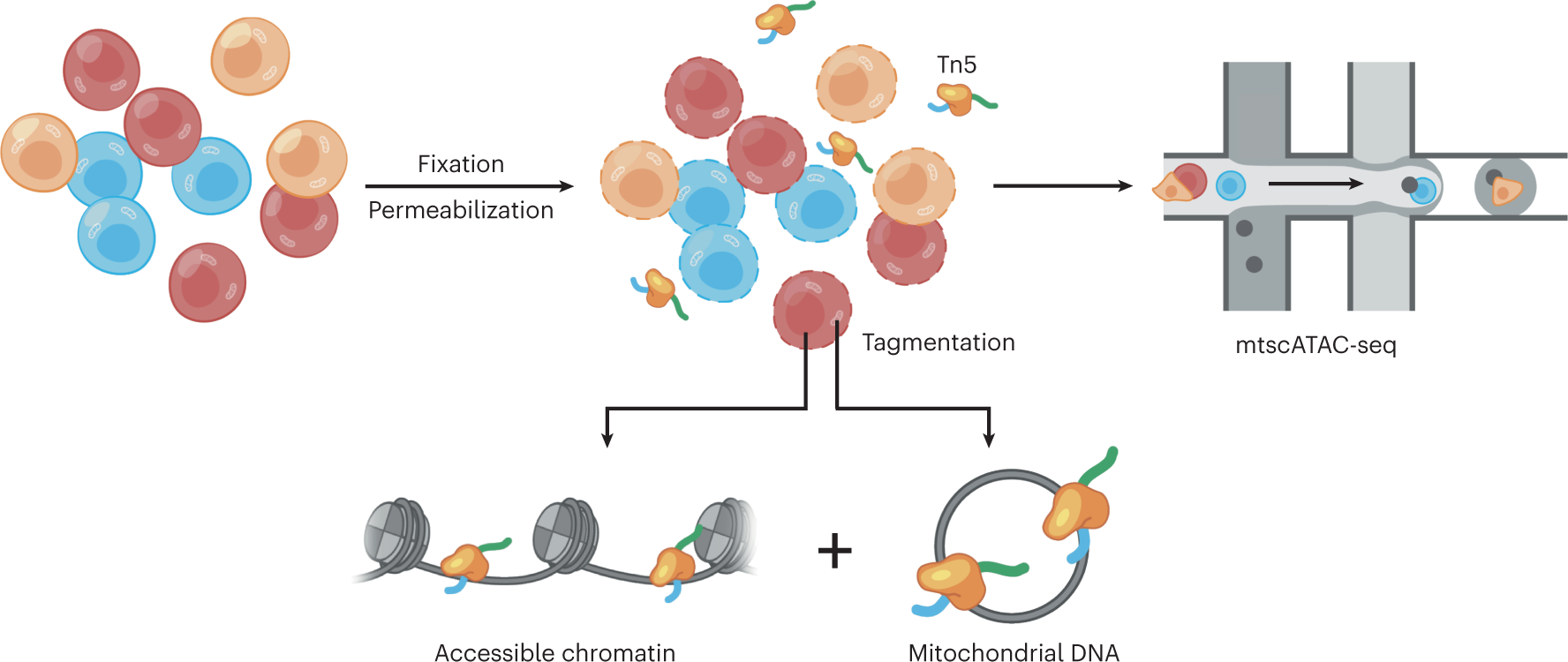
Schematic of the mtscATAC-seq experimental workflow
To retain mtDNA within their host cells, whole cells are fixed before mild lysis and permeabilization. Permeabilization enables access of the Tn5 transposase for transposition of nuclear accessible chromatin and mtDNA. Tagmented cells are encapsulated into droplets by using the Chromium Next GEM Single Cell ATAC platform by 10x Genomics to achieve single-cell compartmentalization and library generation.
Lareau CA, Liu V, Muus C, Praktiknjo SD, Nitsch L, Kautz P, Sandor K, Yin Y, Gutierrez JC, Pelka K, Satpathy AT, Regev A, Sankaran VG, Ludwig LS. (2023) Mitochondrial single-cell ATAC-seq for high-throughput multi-omic detection of mitochondrial genotypes and chromatin accessibility. Nat Protoc [Epub ahead of print]. [abstract]