N6-methyladenosine (m6A), the most abundant internal messenger RNA modification in higher eukaryotes, serves myriad roles in regulating cellular processes. Functional dissection of m6A is, however, hampered in part by the lack of high-resolution and quantitative detection methods. Researchers at the University of Chicago have developed evolved TadA-assisted N6-methyladenosine sequencing (eTAM-seq), an enzyme-assisted sequencing technology that detects and quantifies m6A by global adenosine deamination. With eTAM-seq, they analyze the transcriptome-wide distribution of m6A in HeLa and mouse embryonic stem cells. The enzymatic deamination route employed by eTAM-seq preserves RNA integrity, facilitating m6A detection from limited input samples. In addition to transcriptome-wide m6A profiling, the researchers demonstrate site-specific, deep-sequencing-free m6A quantification with as few as ten cells, an input demand orders of magnitude lower than existing quantitative profiling methods. They envision that eTAM-seq will enable researchers to not only survey the m6A landscape at unprecedented resolution, but also detect m6A at user-specified loci with a simple workflow.
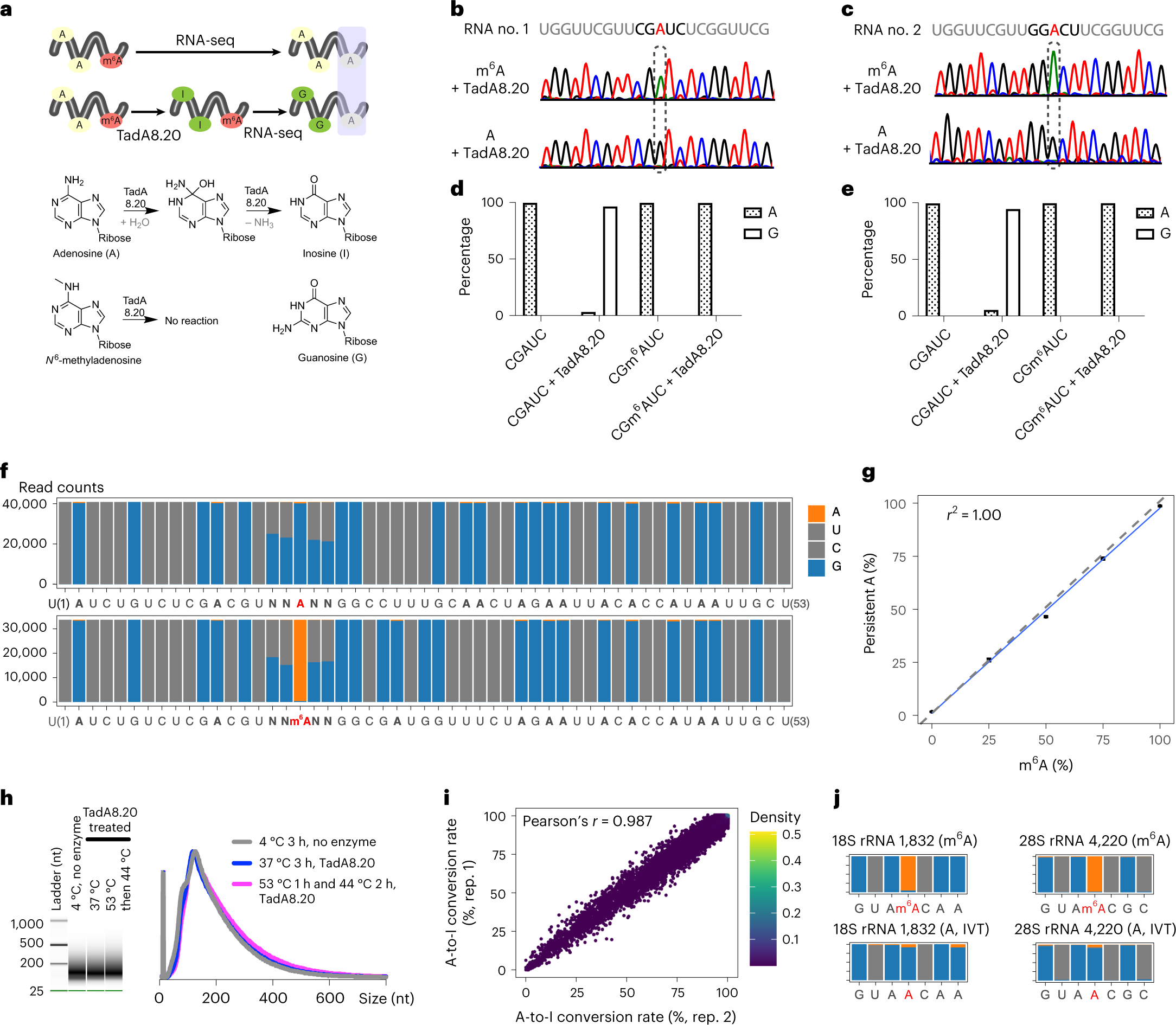
Global A deamination by TadA8.20
a, Proposed m6A detection scheme. TadA8.20 selectively converts A into I, without acting on m6A. I is recognized as G by reverse transcriptases. Persistent A post-TadA8.20 treatment corresponds to m6A. b,c, In vitro deamination of RNA probes hosting A or m6A in ‘CGAUC’ (b) and ‘GGACU’ (c) motifs by TadA8.20. Unmethylated and methylated RNA sequences were prepared through in vitro transcription using ATP and N6-methyl-ATP as starting materials, respectively. Treated RNA was reverse transcribed, amplified and subjected to Sanger sequencing. d,e, TadA8.20-catalyzed A-to-I conversion rates in ‘CGAUC’ (d) and ‘GGACU’ (e) probes quantified by next-generation sequencing (NGS). f, Deamination of synthetic A/m6A RNA probes by TadA8.20. The 53-nt RNA probes hosting NNANN and NNm6ANN motifs were treated by TadA8.20. Deaminated RNA underwent RT and NGS. g, Correlation of persistent A signals captured by eTAM-seq and m6A contents in RNA probes. h, Capillary gel electrophoresis analysis of fragmented HeLa mRNA treated with or without TadA8.20 at different temperatures for 3 h. RNA size distribution is plotted on the right. For eTAM-seq, RNA is incubated with TadA8.20 at 53 °C for 1 h followed by a 2-h treatment at 44 °C. Experiments were repeated independently with similar results. i, Transcriptome-wide A-to-I conversion rates in two independent replicates (rep.). Of A sites with ≥100 counts, 10% were randomly sampled to make the scatter plot. Pearson’s r was calculated for all A sites with ≥100 counts. j, Two m6A sites in human rRNA. Positions 1829–1835 of 18S rRNA and positions 4217–4223 of 28S rRNA are plotted
Xiao YL, Liu S, Ge R, Wu Y, He C, Chen M, Tang W. (2022) Transcriptome-wide profiling and quantification of N6-methyladenosine by enzyme-assisted adenosine deamination. Nat Biotechnol [Epub ahead of print]. [abstract]