Bulk sequencing experiments (single- and multi-omics) are essential for exploring wide-ranging biological questions. To facilitate interactive, exploratory tasks, coupled with the sharing of easily accessible information, University of Cambridge researchers present bulkAnalyseR, a package integrating state-of-the-art approaches using an expression matrix as the starting point (pre-processing functions are available as part of the package). Static summary images are replaced with interactive panels illustrating quality-checking, differential expression analysis (with noise detection) and biological interpretation (enrichment analyses, identification of expression patterns, followed by inference and comparison of regulatory interactions). bulkAnalyseR can handle different modalities, facilitating robust integration and comparison of cis-, trans- and customised regulatory networks.
Workflow and examples of bulkAnalyseR summary plots on bulk mRNAseq data
(A) Workflow diagram of the main components of the pipeline and package. (B) PCA plot of all samples illustrating sample similarity. (C) Volcano plot showcasing DE genes between the 0 and 12h samples. (D) Heatmap of Z-scores across samples for the top differentially expressed genes. (E) Enrichment analysis results for the DE genes (enriched terms ordered are by significance). (F) Cross-plot comparing two DE outputs (log2(FC) from 0 versus 12h on the x-axis and 0 versus 36h on the y-axis).
Moutsopoulos I, Williams EC, Mohorianu II. (2022) bulkAnalyseR: an accessible, interactive pipeline for analysing and sharing bulk multi-modal sequencing data. Brief Bioinform [Epub ahead of print]. [article]

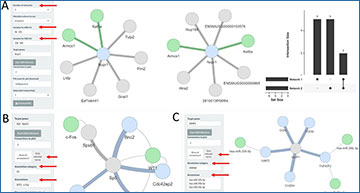
![Workflow and examples of bulkAnalyseR summary plots on bulk mRNAseq data [13]. (A) Workflow diagram of the main components of the pipeline and package; further details are presented in Supplementary Figure S1. (B) PCA plot of all samples illustrating sample similarity. (C) Volcano plot showcasing DE genes between the 0 and 12h samples. (D) Heatmap of Z-scores across samples for the top differentially expressed genes. (E) Enrichment analysis results for the DE genes (enriched terms ordered are by significance). (F) Cross-plot comparing two DE outputs (log2(FC) from 0 versus 12h on the x-axis and 0 versus 36h on the y-axis).](https://i1.wp.com/oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/bib/PAP/10.1093_bib_bbac591/1/bbac591f1.jpeg?Expires=1675775313&Signature=fnyhj8MpQT1jYESKLLaRIXQJClb3FD~vQmzXQbvMptbkJZ9zhDuPqZDe9DRZacD2-LhLhtzdJE~1YzCtwNhDAW~XztDQAedD2V91OclTvmZpweK7UGR2Kh3wF855IaBWtPciQIgOxNFnNQ2JcnDxkuspihCRic9B2pYWvRTeHWLIZj8XDmtOI7F0tHN2PYcN6SG06ukDwwLO6TFa3-Nk3Ipy9aKERIPD6hcDBeRHIVCf~iLcNhewv1wzd3GDh~m1YUY-mZXiDdLpWks5x4zG~W~gQ~Yc3pQrJpD8UnAw3jG~6Q2YJA35NGqciWSjY83AXCXodzVbg8OMKOieVJdVPg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


