RNA molecules can form secondary and tertiary structures that can regulate their localization and function. Using enzymatic or chemical probing together with high-throughput sequencing, secondary structure can be mapped across the entire transcriptome. However, a limiting factor is that only population averages can be obtained since each read is an independent measurement. Although long-read sequencing has recently been used to determine RNA structure, these methods still used aggregate signals across the strands to detect structure. Averaging across the population also means that only limited information about structural heterogeneity across molecules or dependencies within each molecule can be obtained.
University of Bergen researchers have developed Single-Molecule Structure sequencing (SMS-seq) that combines structural probing with native RNA sequencing to provide non-amplified, structural profiles of individual molecules with novel analysis methods. This new approach using mutual information enabled single molecule structural interrogation. Each RNA is probed at numerous bases enabling the discovery of dependencies and heterogeneity of structural features. The researchers also show that SMS-seq can capture tertiary interactions, dynamics of riboswitch ligand binding, and mRNA structural features.
Oxford nanopore direct RNA structure profiling of a hairpin RNA
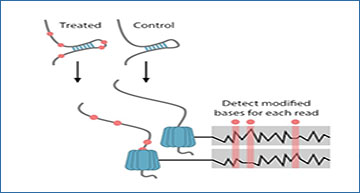
(A) Molecular structure of Adenosine modification by DEPC that modifies structurally unconstrained adenosine by opening the imidazole ring. (B) SMS-seq workflow. Structures are probed with DEPC and the current signal of each nucleotide that passes through the pore is recorded. Modified sites (red) are detected by comparing the current signal from treated samples to the control samples. (C) Normalized current mean and standard deviation for a nucleotide at a single-stranded region of RNA modified with DEPC (red) and unmodified (gray). (D) ROC curves for unmodified, modified, and denatured 5-mer RNA sequences. The 5-mer RNAs are treated with 1% (black), 5% (blue), 10% (orange) DEPC as well as denatured RNA treated with 5% DEPC (green). (E) Denaturing 15% polyacrylamide gel electrophoresis of 5′-end radiolabeled RNA fragments generated from DEPC treated hairpin RNA (top) and untreated hairpin RNA (bottom), which were treated with borohydride and aniline. Arrows mark positions that were likely to be identified by SMS-seq as modified. (F) Proton NMR spectra of non-treated (blue) and DEPC treated (red) for hairpin RNA. Singlet of protons on positions 8 and 2 on the purine ring could be quantified. (G) Tombo modification statistics for adenosine bases. Bases are called modified (open) when their statistic is below 0.2 (dotted line) corresponding to an FDR of 15%. (H) Heatmap showing 3093 full-length reads (y-axis) with modified (red) and unmodified (gray) nucleotides at each position (x-axis) for each read. Reads are ordered by the overall modification rate. The legend above (Structure) shows which nucleotides are accessible (white) and inaccessible (blue) in the hairpin structure. (I) Modification frequency per base over the hairpin structure. Position 29 is uncharacteristic of a stem and is called as modified to a greater degree. This high modification rate could be due to RNA conformational heterogeneity, RNA breathing, or a problematic k-mer. (J) ROC curve for the hairpin at the consensus level (black) and individual bases (orange). A random model is shown with the dotted line. (K) Groups of reads folding to the same structure are shown by abundance (y-axis) and minimum folding energy (x-axis). The inset contains selected structures with modified bases colored (red circles).
Bizuayehu TT, Labun K, Jakubec M, Jefimov K, Niazi AM, Valen E. (2022) Long-read single-molecule RNA structure sequencing using nanopore. Nucleic Acids Res [Epub ahead of print]. [article]