Recently, single-cell RNA sequencing (scRNA-seq) and single-cell assay for transposase-accessible chromatin using sequencing (scATAC-seq) have been developed to separately measure transcriptomes and chromatin accessibility profiles at the single-cell resolution. However, few methods can reliably integrate these data to perform regulatory network analysis.
Researchers at the Guangzhou Institutes of Biomedicine and Health have developed integrated regulatory network analysis (IReNA) for network inference through the integrated analysis of scRNA-seq and scATAC-seq data, network modularization, transcription factor enrichment, and construction of simplified intermodular regulatory networks. Using public datasets, the researchers showed that integrated network analysis of scRNA-seq data with scATAC-seq data is more precise to identify known regulators than scRNA-seq data analysis alone. Moreover, IReNA outperformed currently available methods in identifying known regulators. IReNA facilitates the systems-level understanding of biological regulatory mechanisms.
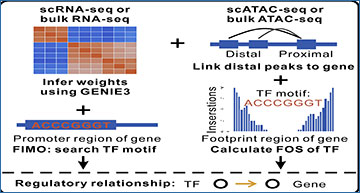
Flowchart of IReNA
IReNA consists of two components: network inference and network decoding. Network inference is performed according to weights calculated by GENIE3. FIMO is then used to identify binding motifs of transcription factors and refine regulatory relationships if only gene expression profiles (single-cell or bulk RNA-seq data) are used. If bulk or single-cell ATAC-seq data are available, binding motifs and footprint occupancy score (FOS) are used to refine regulatory relationships inferred from gene expression analysis. Especially for single-cell ATAC-seq data, ArchR is used to link peaks and genes. Next, inferred networks are modularized and used to enrich transcription factors (TFs). Regulatory networks of enriched TFs are further decoded by establishing simplified regulatory networks among modules.
Availability – IReNA is available at https://github.com/jiang-junyao/IReNA.
Jiang J, Lyu P, Li J, Huang S, Tao J, Blackshaw S, Qian J, Wang J. (2022) IReNA: Integrated regulatory network analysis of single-cell transcriptomes and chromatin accessibility profiles. iScience 25(11):105359. [article]