Alternative polyadenylation (APA) is a major mechanism that increases transcriptional diversity and regulates mRNA abundance. Existing computational tools to analyze APA have low precision because these tools are designed for short-read RNA-seq, which is a suboptimal data source to study APA. Long-read RNA-seq (LR-RNA-seq) accurately detects complete transcript isoforms with poly(A)-tails, providing an ideal data source to study APA. However, current computational tools are incompatible with LR-RNA-seq.
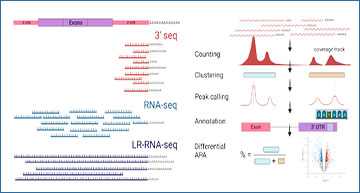
Researchers from the University of California, Irvine have developed LAPA, a computational toolkit to study alternative polyadenylation (APA) from diverse data sources such as LR-RNA-seq and short-read 3′ sequencing (3′-seq). LAPA counts and clusters reads with poly(A)-tail, then performs peak-calling to detect poly(A)-site in a data source agnostic manner. The resulting peaks are annotated based on genomics features and regulatory sequence elements such as presence of a poly(A)-signal. Finally, LAPA can perform robust statistical testing and multiple testing correction to detect differential APA.
Computational steps of LAPA
The researchers analyzed ENCODE LR-RNA-seq data from human WTC11, mouse C2C12 myoblast, and C2C12-derived differentiated myotube cells using LAPA. Comparing LR-RNA-seq from different platforms and library preparation methods against 3′-seq shows that LR-RNA-seq detects poly(A)-sites with a performance of 75% precision at 57% recall. Moreover, LAPA consistently improved TES validation by at least 25% over the baseline transcriptome annotation generated by TALON, independent of protocol or platform. Differential APA analysis detected 788 statistically significant genes with unique polyadenylation signatures between undifferentiated myoblast and differentiated myotube cells. Among these genes, 3′ UTR elongation is significantly associated with higher expression, while shortening is linked with lower expression. This analysis reveals a link between cell state/identity and APA. Overall, these results show that LR-RNA-seq is a reliable data source for the study of post-transcriptional regu- lation by providing precise information about alternative polyadenylation.
Availability: LAPA is publicly available at https://github.com/mortazavilab/lapa and PyPI.
Celik MH, Mortazavi A. (2022) Analysis of alternative polyadenylation from long-read or short-read RNA-seq with LAPA. bioRXiv [online pre-print]. [abstract]