Single-cell RNA sequencing (scRNA-seq) data contain rich information at the gene, transcript, and nucleotide levels. Most analyses of scRNA-seq have focused on gene expression profiles, and it remains challenging to extract nucleotide variants and isoform-specific information. Researchers at UCLA have developed scAllele, an integrative approach that detects single-nucleotide variants, insertions, deletions, and their allelic linkage with splicing patterns in scRNA-seq. The researchers demonstrate that scAllele achieves better performance in identifying nucleotide variants than other commonly used tools. In addition, the read-specific variant calls by scAllele enables allele-specific splicing analysis, a unique feature not afforded by other methods. Applied to a lung cancer scRNA-seq dataset, scAllele identified variants with strong allelic linkage to alternative splicing, some of which are cancer specific and enriched in cancer-relevant pathways. scAllele represents a versatile tool to uncover multilayer information and previously unidentified biological insights from scRNA-seq data.
Algorithm outline
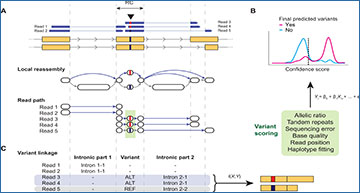
(A) Illustration of the main algorithm of scAllele for variant calling. The reads and the reference genomic sequence overlapping an RC are decomposed into k-mers and reassembled into a de Bruijn graph. The graph shown here is a compacted version. The bubbles in the graph indicate a sequence mismatch, i.e., a variant. For each read, scAllele obtains a path for the original read sequence and infers the allele of each variant (including introns). (B) Variants (green box in A) identified from the graph are then scored using a GLM. The GLM was trained with different features (green box) to assign a confidence score to the variants. See Materials and Methods for details. (C) To identify allele-specific splicing (i.e., variant linkage), scAllele performs a mutual information (MI) calculation between nucleotide variants (SNVs and microindels) and intronic parts (where the alleles are the different overlapping introns) to calculate allelic linkage of splicing isoforms
Quinones-Valdez G, Fu T, Chan TW, Xiao X. (2022) scAllele: A versatile tool for the detection and analysis of variants in scRNA-seq. Sci Adv 8(35):eabn6398. [article]