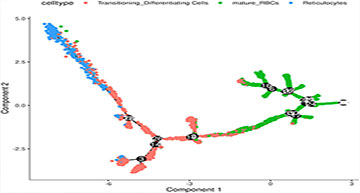
Human red blood cells (RBCs), or erythrocytes, are the most abundant blood cells responsible for gas exchange. RBC diseases affect hundreds of millions of people and impose enormous financial and personal burdens. One well-recognized, but poorly understood feature of RBC populations within the same individual are their phenotypic heterogeneity. The granular characterization of phenotypic RBC variation in normative and disease states may allow us to identify the genetic determinants of red cell diseases and reveal novel therapeutic approaches for their treatment. Previously, researchers at the Duke Molecular Physiology Institute discovered diverse RNA transcripts in RBCs that has allowed them to dissect the phenotypic heterogeneity and malaria resistance of sickle red cells. However, these analyses failed to capture the heterogeneity found in RBC sub-populations. To overcome this limitation, the researchers have performed single cell RNA-Seq to analyze the transcriptional heterogeneity of RBCs from three adult healthy donors which have been stored in the blood bank conditions and assayed at day 1 and day 15. The expression pattern clearly separated RBCs into seven distinct clusters that include one RBC cluster that expresses HBG2 and a small population of RBCs that express fetal hemoglobin (HbF) that they annotated as F cells. Almost all HBG2-expessing cells also express HBB, suggesting bi-allelic expression in single RBC from the HBG2/HBB loci, and we annotated another cluster as reticulocytes based on canonical gene expression. Additional RBC clusters were also annotated based on the enriched expression of NIX, ACVR2B and HEMGN, previously shown to be involved in erythropoiesis. Finally, the researchers found the storage of RBC was associated with an increase in the ACVR2B and F-cell clusters. Collectively, these data indicate the power of single RBC RNA-Seq to capture and discover known and unexpected heterogeneity of RBC population.
The relationship between the RBC clusters and storage times
(A) The indicated storage days of individual RBCs were projected into the UMAP clusters of RBCs. (B) The UMAP presentation of the heterogeneous clusters of separate RBCs which have been stored for one (left) or 15 days (right). (C) The relative frequency of the seven representative RBC clusters in day 1 vs. 15 showing an increase in ACVR2B and F-cells in ex vivo stored RBCs.
Jain V, Yang WH, Wu J, Roback JD, Gregory SG, Chi JT. (2022) Single Cell RNA-Seq Analysis of Human Red Cells. Front Physiol 13:828700. [article]