Metagenomics and total RNA sequencing (total RNA-Seq) have the potential to improve the taxonomic identification of diverse microbial communities, which could allow for the incorporation of microbes into routine ecological assessments. However, these target-PCR-free techniques require more testing and optimization. University of Guelph researchers processed metagenomics and total RNA-Seq data from a commercially available microbial mock community using 672 data-processing workflows, identified the most accurate data-processing tools, and compared their microbial identification accuracy at equal and increasing sequencing depths. The accuracy of data-processing tools substantially varied among replicates. Total RNA-Seq was more accurate than metagenomics at equal sequencing depths and even at sequencing depths almost one order of magnitude lower than those of metagenomics. The researchers show that while data-processing tools require further exploration, total RNA-Seq might be a favorable alternative to metagenomics for target-PCR-free taxonomic identifications of microbial communities and might enable a substantial reduction in sequencing costs while maintaining accuracy. This could be particularly an advantage for routine ecological assessments, which require cost-effective yet accurate methods, and might allow for the incorporation of microbes into ecological assessments.
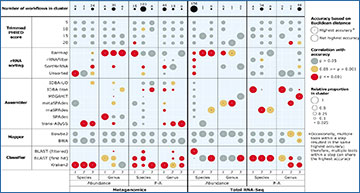
Relative frequency of data-processing tools within clusters of most accurate workflows (circle size), significance of correlations between tools and accuracy (circle colour), and most accurate tools based on different evaluation levels (dot in circle centre). Evaluation levels consisted of combinations of sequencing type (metagenomics/total RNA-Seq), data type (abundance/P–A), and taxonomic rank (genus/species). Each column represents one evaluation level utilized for one of three replicates. The relative frequency of tools and the tool with the highest accuracy were determined for each data-processing step separately. Performances differed substantially among replicates and evaluation levels.
Hempel CA, Wright N, Harvie J, Hleap JS, Adamowicz SJ, Steinke D. (2022) Metagenomics versus total RNA sequencing: most accurate data-processing tools, microbial identification accuracy and perspectives for ecological assessments. Nucleic Acids Res [Epub ahead of print]. [article]