Broad Institute researchers have developed BacDrop, a highly scalable technology for bacterial single-cell RNA sequencing that has overcome many challenges hindering the development of scRNA-seq in bacteria. BacDrop can be applied to thousands or millions of cells from both gram-negative and gram-positive species. It features universal ribosomal RNA depletion and combinatorial barcodes that enable multiplexing and massively parallel sequencing. The researchers applied BacDrop to study Klebsiella pneumoniae clinical isolates and to elucidate their heterogeneous responses to antibiotic stress. In an unperturbed population presumed to be homogenous, they found within-population heterogeneity largely driven by the expression of mobile genetic elements that promote the evolution of antibiotic resistance. Under antibiotic perturbation, BacDrop revealed transcriptionally distinct subpopulations associated with different phenotypic outcomes including antibiotic persistence. BacDrop thus can capture cellular states that cannot be detected by bulk RNA-seq, which will unlock new microbiological insights into bacterial responses to perturbations and larger bacterial communities such as the microbiome.
BacDrop: a bacterial droplet-based massively parallel scRNA-seq technology
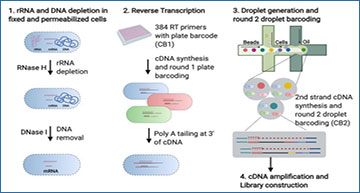
(A) BacDrop workflow. Following cell fixation and permeabilization, rRNA and gDNA is depleted from cells in bulk. Then CB1 and UMIs are added to the 5’ end of cDNA via RT reactions (round 1 plate barcoding) in pools of ~200,000 cells per well in 96- or 384-well plates. After round 1 plate barcoding, all cells are pooled and cDNA is polyadenylated at the 3’ end using terminal transferase, followed by droplet generation and round 2 droplet barcoding. The 3’ poly-A tail of cDNA enables second strand synthesis using oligo-dT primers. Round 2 droplet barcoding is achieved via second strand cDNA synthesis and 4 capturing cycles by barcoded primers (on 10x gel beads) in droplets. The successfully captured cDNA contains UMIs, CB1 and CB2, as well as adaptor sequences at both 5’ and 3’ end. Each cell is identified by a combination of CB1 and CB2. (B) rRNA depletion inside fixed and permeabilized cells significantly increases the percentage of total reads that align to mRNA in BacDrop libraries of K. pneumoniae. (C) rRNA depletion does not affect mRNA profiles. BacDrop libraries were constructed using in K. pneumoniae samples with or without rRNA depletion, and a linear regression model was fitted to the mRNA counts from each library. (D) Cell fixation and permeabilization does not affect mRNA profiles. Bulk RNA-seq results derived from Trizol-extracted RNA samples of K. pneumoniae versus RNA derived from fixed and permeabilized K. pneumoniae cells were highly correlated. (E) Single-cell BacDrop results are highly correlated with bulk RNA-seq results when analyzed in bulk mode (without cell barcode extraction). (F) BacDrop has low barcode collision rates in an experiment where 2 million bacterial cells, mixed with K. pneumoniae and P. aeruginosa cells, were loaded into one 10x channel (~6 cells per droplet). About 2.8 % of the cells was assigned to two species, resulting in a 6.6% barcode collision rate. (G) BacDrop was performed on 4 different bacterial species including the gram-negative E. coli, K. pneumoniae, P. aeruginosa, and the gram-positive bacterium E. faecium. Uniform Manifold Approximation and Projection (UMAP) of this mixed population shows the separation of different species, colored by species identity.
Ma P, Amemiya HM, He LL, Gandhi SJ, Nicol R, Bhattacharyya RP, Smillie CS, Hung DT. (2022) Bacterial droplet-based single-cell RNA-seq reveals heterogeneity in bacterial populations and in response to antibiotic perturbation. bioRXiv [online preprint]. [abtract]