Recent advances in biochemistry and single-cell RNA sequencing (scRNA-seq) have allowed us to monitor the biological systems at the single-cell resolution. However, the low capture of mRNA material within individual cells often leads to inaccurate quantification of genetic material. Consequently, a significant amount of expression values are reported as missing, which are often referred to as dropouts.
To overcome this challenge, researchers at the University of Nevada Reno have developed a novel imputation method, named single-cell Imputation via Subspace Regression (scISR), that can reliably recover the dropout values of scRNA-seq data. The scISR method first uses a hypothesis-testing technique to identify zero-valued entries that are most likely affected by dropout events and then estimates the dropout values using a subspace regression model. The researchers comprehensive evaluation using 25 publicly available scRNA-seq datasets and various simulation scenarios against five state-of-the-art methods demonstrates that scISR is better than other imputation methods in recovering scRNA-seq expression profiles via imputation. scISR consistently improves the quality of cluster analysis regardless of dropout rates, normalization techniques, and quantification schemes.
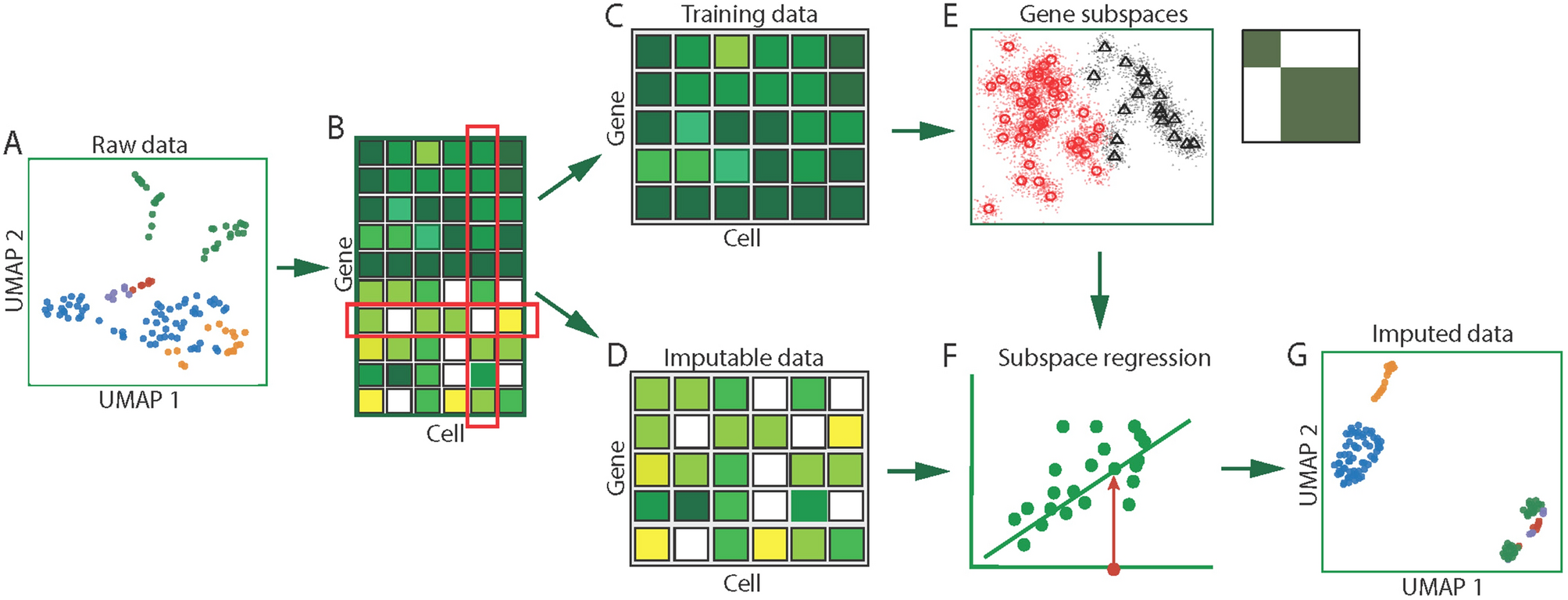
Single-cell Imputation using Subspace Regression (scISR)
(A) Input data visualized in cell/sample space. (B) Hypergeometric test to determine whether each zero value is induced by dropout. Based on the computed p-values for each entry, we separate the original data into two sets of data: training data and imputable data. (C) Training data in which none of the values is induced by dropout events. (D) Imputable data in which each gene has at least one entry that is likely to be induced by dropout events. (E) Gene subspaces determined by perturbation clustering. We perturb the training data to discover the natural structure of the genes. Based on the pair-wise similarity between genes, we separate genes into groups that share similar patterns. (F) Subspace regression. We assign each gene in the imputable data to the closest subspace and then perform a generalized linear regression on the subspace to estimate the zero-valued entries that are impacted by dropouts. (G) Output expression matrix obtained by concatenating the training data and imputed data.
Availability – The source code of scISR can be found on GitHub at: https://github.com/duct317/scISR
Tran D, Tran B, Nguyen H, Nguyen T. (2022) A novel method for single-cell data imputation using subspace regression. Sci Rep 12(1):2697. [article]