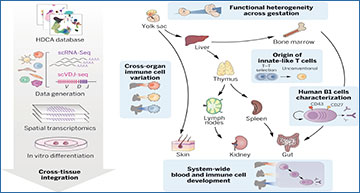
The human immune system develops across several anatomical sites throughout gestation. Immune cells differentiate initially from extra-embryonic yolk sac progenitors and subsequently from aorto-gonad-mesonephros–derived hematopoietic stem cells before liver and bone marrow take over as the primary sites of hematopoiesis. Immune cells from these primary hematopoietic sites then seed developing lymphoid organs and peripheral nonlymphoid organs. Recent advances in single-cell genomics technologies have facilitated studies on the developing immune system at unprecedented scale and resolution. However, these studies have focused on one or a few organs rather than reconstructing the entire immune system as a distributed network across tissues.
To provide a detailed characterization of the developing immune system across multiple organs, Wellcome Sanger Institute researchers performed single-cell RNA sequencing (scRNA-seq) using dissociated cells from yolk sac, prenatal spleen, and skin, and integrated publicly available cell atlases of six additional organs, spanning weeks 4 to 17 after conception. To further characterize developmental B and T cells and explore their antigen receptor repertoire, they also generated paired γδ T cell receptor (γδTCR)–, αβ T cell receptor (αβTCR)–, and B cell receptor (BCR)–sequencing data. Finally, to study the spatial localizations of cell populations in early hematopoietic tissue and lymphoid organs critical for B and T cell development, the researchers performed spatial transcriptomics on prenatal spleen, liver, and thymus and used the scRNA-seq data as a reference to map the cells in situ.
These researchers have integrated a cross-tissue single-cell atlas of developing human immune cells across prenatal hematopoietic, lymphoid, and nonlymphoid peripheral organs. This includes over 900,000 cells from which they identified over 100 cell states.
Using cross-gestation analysis, the researchers revealed the acquisition of immune-effector functions of myeloid and lymphoid cell types from the second trimester, and their early transcriptomic signatures suggested a role in tissue morphogenesis. Through cross-organ analysis, they identified conserved processes of proliferation and maturation for monocytes and T cells before their migration from the bone marrow and thymus, respectively, into peripheral tissues. They discovered system-wide blood and immune cell development, in particular B lymphopoiesis, across all sampled peripheral organs. This expands on previous understanding of conventional hematopoietic organs (yolk sac, liver, and bone marrow) as the only sites for immune cell development. They validated the presence and location of lineage-committed progenitors spatially using 10X Genomics Visium Spatial Gene Expression and single-molecule fluorescence in situ hybridization. Finally, the researchers identified and functionally validated the properties of human prenatal innate-like B and T cells, providing an extensive characterization of human B1 cells with single-cell transcriptomic and BCR information, as well as functional validation of spontaneous antibody secretion. Integrating the transcriptome profiles of human prenatal unconventional T cells, their αβTCR V(D)J usage, and data from an in vitro thymic organoid culture model, they supply additional evidence for thymocyte–thymocyte selection during unconventional T cell development.
Cross-tissue mapping of the developing human immune system
We reconstructed the process of immune cell development, analyzing cells across prenatal hematopoietic, lymphoid, and peripheral organs, combining scRNA-seq, scVDJ-seq, and spatial transcriptomics. With this integrated dataset, we studied variation in cellular phenotypes across development and between tissues and the distribution of blood and immune cell progenitors across tissues and characterized fetal-specific innate-like B and T cells.
This comprehensive single-cell and spatial atlas of the developing human immune system provides valuable resources and biological insights to facilitate in vitro cell engineering and regenerative medicine and to enhance our understanding of congenital disorders affecting the immune system.
Suo C, Dann E, Goh I, Jardine L, Kleshchevnikov V, Park JE, Botting RA, Stephenson E, Engelbert J, Tuong ZK, Polanski K, Yayon N, Xu C, Suchanek O, Elmentaite R, Domínguez Conde C, He P, Pritchard S, Miah M, Moldovan C, Steemers AS, Mazin P, Prete M, Horsfall D, Marioni JC, Clatworthy MR, Haniffa M, Teichmann SA. (2022) Mapping the developing human immune system across organs. Science 376(6597):eabo0510. [abstract]