The creation and analysis of gene regulatory networks have been the focus of bioinformatics research and underpins much of what is known about gene regulation. However, as a result of a bias in the availability of data-types that are collected, the vast majority of gene regulatory network resources and tools have focused on either transcriptional regulation or protein-protein interactions. This has left other areas of regulation, for instance, translational regulation, vastly underrepresented despite them having been shown to play a critical role in both health and disease.
In order to address this researchers at Duke-NUS Medical School have developed CLIPreg, a package that integrates RNA, Ribo and CLIP- sequencing data in order to construct translational regulatory networks coordinated by RNA-binding proteins and micro-RNAs. This is the first tool of its type to be created, allowing for detailed investigation into a previously unseen layer of regulation.
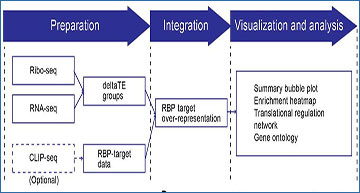
(a) CLIPreg workflow: Step 1, Data preparation: Ribo-seq and RNA-seq data are used to determine translational efficiency changes using deltaTE and CLIP-seq data is used to determine RBP-target relations. Step 2, Data integration: RNA-binding protein (RBP)-target over-representation test to identify key RBPs regulating translation. Step 3, Data visualisation: Visualisation and analysis. (b) Heatmap of RBP enrichment per gene group. -log(P) denotes the false discovery rate. RBP Log2FC denotes RBP’s Ribo-seq log2 fold change. Orange: RBP upregulated; Blue: RBP downregulated. Quaking (QKI) and Pumillo-2 (PUM2) highlighted for detailed view. (c) Bubble plot showing RBP enrichment per regulation group. (d) RBP Translational regulation network. Red: RBP; Blue: Targets with upregulated translational-efficiency (TE); Orange: Targets with downregulated TE. (e) Gene ontology of RBP regulated gene sets as labelled in the network. ID2 and ID16 groups shown as examples.
Availability: CLIPreg is available at https://github.com/SGDDNB/CLIPreg.
Kerouanton B, Schäfer S, Ho L, Chothani S, Rackham OJL. (2022) CLIPreg: Constructing translational regulatory networks from CLIP-, Ribo- and RNA-seq. Bioinformatics [Epub ahead of print]. [abstract]