Single-cell RNA sequencing (scRNA-seq) is a groundbreaking technique that allows scientists to explore gene expression in individual cells within a complex sample. This technology can reveal the diversity and behavior of cells in unprecedented detail, making it a vital tool for many areas of biological research, from understanding disease mechanisms to developing new treatments. However, the complexity of analyzing scRNA-seq data often requires advanced mathematical and programming skills, which can be a barrier for many research teams.
To bridge this gap between laboratory-based (wet-lab) researchers and data analysis (dry-lab) experts, researchers at the Universidad Autónoma de Madrid have developed a new tool called SinglePointRNA. Here’s how SinglePointRNA makes single-cell transcriptomics more accessible to researchers, regardless of their computational background.
What is SinglePointRNA?
SinglePointRNA is an application based on the R programming language and designed with a user-friendly graphical interface using the Shiny framework. It integrates several publicly available tools for analyzing single-cell RNA-seq data, offering a comprehensive suite of features that guide researchers through the complex data analysis process.
SinglePointRNA implementation overview
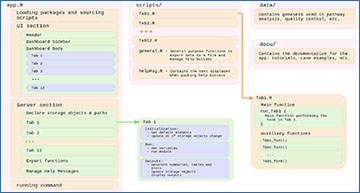
SinglePointRNA is composed of four main sections. The script “app.R” contains the core of the program. Structured in two blocks, “UI” and “server”, as most shiny apps are, “app.R” sets the interface elements of each tab, as well as handles the inputs and outputs of each task. The “scripts/” folder contains all the code needed to perform each task, with one self-contained script per tab. The “data/” folder holds gene set lists and gene naming patterns used for pathway analysis and quality control, stored in plain text files so they can be easily expanded. Finally, the “docu/” folder stores the documentation displayed in the first tab of SinglePointRNA: the user guide, tutorials, etc.
Key Features and Benefits
- User-Friendly Interface:
- SinglePointRNA provides an intuitive graphical interface that simplifies the analysis process, making it accessible even to those with limited programming experience. This means that researchers can focus more on their scientific questions rather than the technical details of data analysis.
- Comprehensive Analysis Tools:
- The application offers a range of tools for different aspects of scRNA-seq data analysis, including quality control, data normalization, clustering, and visualization. By integrating these tools into a single platform, SinglePointRNA streamlines the workflow, making it easier to perform detailed and customized analyses.
- Context-Driven Framework:
- SinglePointRNA is structured to provide qualitative guidance at each step of the analysis process. This helps users understand what they are doing and why, which is crucial for interpreting the results correctly and making informed decisions about their research.
- Educational Resources:
- To further support researchers, SinglePointRNA includes rich user guides and tutorials. These resources not only help users navigate the software but also serve as an entry point for learning more about the computational techniques used in single-cell data analysis.
- Availability and Accessibility:
- SinglePointRNA and its accompanying datasets for tutorials are freely available on GitHub (www.github.com/ScienceParkMadrid/SinglePointRNA). This open-access approach ensures that a wide range of researchers can benefit from this powerful tool.
Why SinglePointRNA Matters
The development of SinglePointRNA is a significant step towards democratizing access to advanced single-cell RNA sequencing analysis. By providing a tool that combines powerful analysis capabilities with ease of use, SinglePointRNA empowers more researchers to harness the full potential of scRNA-seq data. This can accelerate discoveries in various fields, from cancer research to neurobiology, where understanding the behavior and interaction of individual cells is crucial.
In summary, SinglePointRNA breaks down the barriers between wet-lab and dry-lab research, enabling scientists to perform sophisticated analyses with confidence and ease. By making advanced computational tools accessible to a broader audience, SinglePointRNA holds the promise of advancing scientific research and innovation in single-cell biology.
Puente-Santamaría L, Del Peso L. (2024) SinglePointRNA, an user-friendly application implementing single cell RNA-seq analysis software. PLoS One 19(6):e0300567. [article]