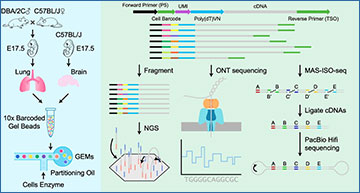
In the ever-evolving field of genetics, scientists are continuously developing new technologies to understand the intricate details of cellular functions. One such breakthrough is single-cell RNA sequencing (scRNA-seq), which allows researchers to detect and measure gene expression at an individual cell level. This technology has revolutionized our ability to study the roles of genes in various biological processes. However, traditional scRNA-seq using next-generation sequencing (NGS) has its limitations. This blog post explores how third-generation sequencing (TGS) technologies, such as Oxford Nanopore Technologies (ONT) and Pacific Biosciences (PacBio), are being used to overcome these limitations and provide a deeper understanding of cellular function.
The Power and Limitations of NGS-based scRNA-seq
Next-generation sequencing (NGS) has made significant strides in high-throughput gene expression analysis. It allows scientists to see which genes are active in a cell and to what extent. However, NGS has a critical limitation: it cannot provide detailed information about the exact structures of the gene transcripts, known as isoforms, due to its relatively short read lengths. Isoforms are different versions of mRNA produced from the same gene, and they can have distinct functions. Therefore, understanding these isoforms is crucial for a comprehensive understanding of gene function.
The Promise of Third-Generation Sequencing
Third-generation sequencing (TGS) technologies, including Oxford Nanopore Technologies (ONT) and Pacific Biosciences (PacBio), offer a solution to the limitations of NGS. These technologies can read much longer sequences of DNA or RNA, allowing scientists to see entire cDNA molecules in one go. This capability makes it possible to identify the exact isoforms of each gene, providing a more detailed picture of gene expression.
Comparing ONT and PacBio in Single-Cell Analyses
To understand how well ONT and PacBio perform in single-cell analyses, researchers at Sun Yat-sen University conducted a study where they generated data from both platforms using the same single-cell cDNA libraries. They then compared the results to those obtained from NGS-based scRNA-seq to assess the performance of each TGS platform in identifying cell types and novel isoforms, as well as allele-specific gene/isoform expression (differences in gene expression depending on the allele).
Key Findings
- Cell Type Identification: Both ONT and PacBio were evaluated for their ability to identify different cell types. Using NGS as a control, researchers found that both TGS platforms could effectively identify cell types from single-cell samples.
- Novel Isoform Detection: One of the significant advantages of TGS is its ability to identify novel isoforms. The study found that PacBio, due to its higher sequencing quality, outperformed ONT in accurately identifying these novel isoforms.
- Allele-Specific Expression: Both platforms were also tested for their ability to detect allele-specific gene and isoform expression. Again, PacBio showed higher accuracy in identifying these subtle genetic differences compared to ONT.
Conclusion
While NGS-based scRNA-seq is excellent for quantifying gene expression, it falls short in revealing the complete picture of transcript structures due to its limited read length. Third-generation sequencing technologies like ONT and PacBio overcome this limitation by providing much longer reads, allowing for detailed isoform analysis and more accurate detection of allele-specific expression. Among the TGS platforms, PacBio stands out for its superior sequencing quality, making it the preferred choice for high-accuracy single-cell transcriptome studies.
This study highlights the potential of TGS technologies to complement and enhance our understanding of gene expression at the single-cell level, paving the way for more precise and comprehensive cellular analysis in research and clinical applications. As these technologies continue to advance, they hold the promise of unlocking new insights into the complex world of cellular functions and disease mechanisms.
Deng E, Shen Q, Zhang J, Fang Y, Chang L, Luo G, Fan X. (2024) Systematic evaluation of single-cell RNA-seq analyses performance based on long-read sequencing platforms. J Adv Res [Epub ahead of print]. [article]