COVID-19 has been a major global health challenge, affecting millions of people worldwide. One of the key strategies to manage the pandemic effectively is to predict the outcomes for patients infected with the SARS-CoV-2 virus. Accurate predictions can help healthcare providers tailor treatments to individual patients, potentially reducing the severity and mortality associated with the disease. In a recent collaborative study involving 15 institutions across Europe, researchers have developed a promising machine learning model that predicts the risk of in-hospital mortality for COVID-19 patients.
The Study: A Collaborative Effort
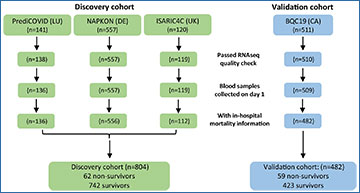
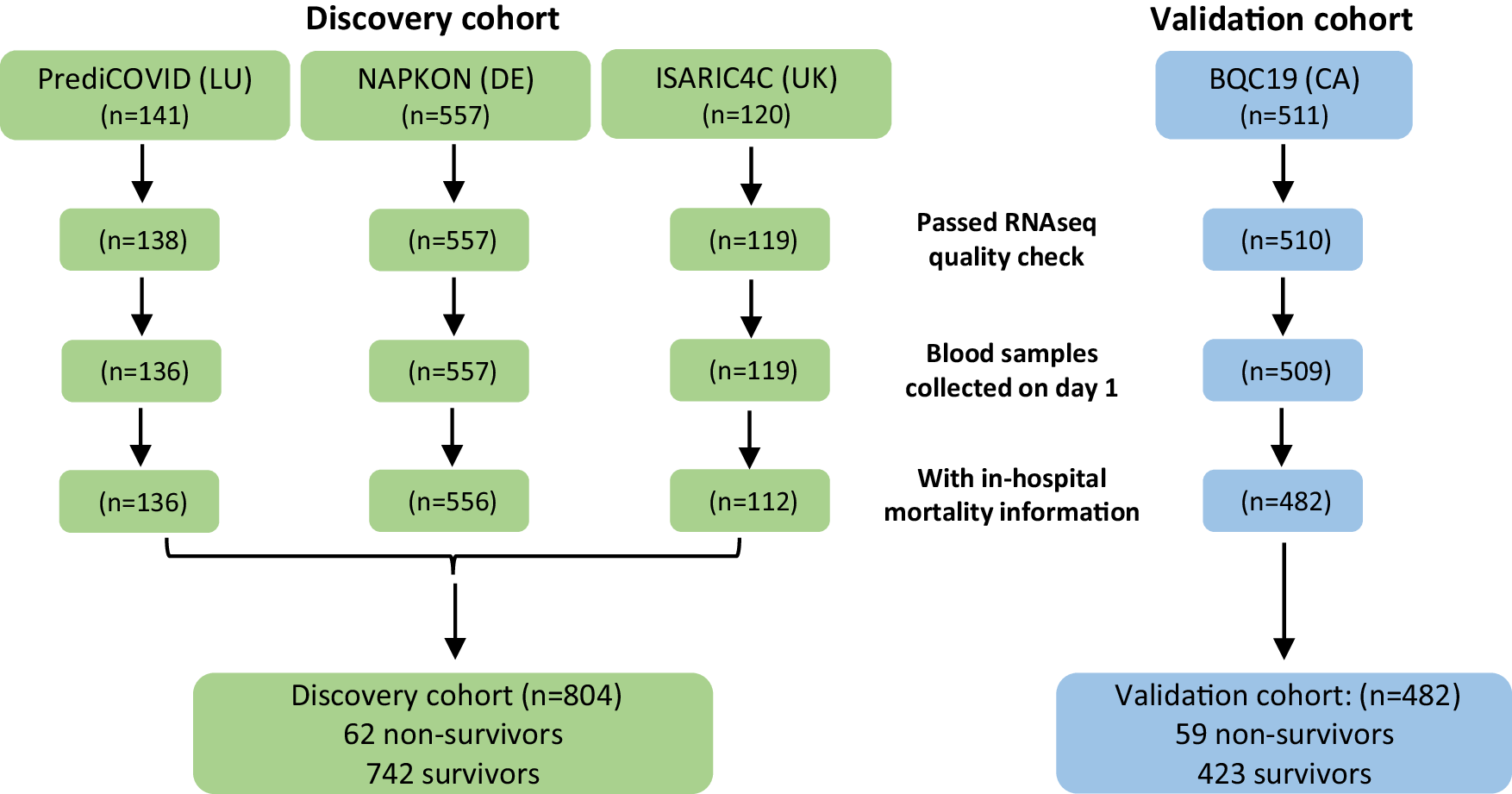
The study analyzed blood samples and clinical data from 1,286 COVID-19 patients collected between 2020 and 2023. These patients were part of four different cohorts from Europe and Canada. The researchers focused on profiling 2,906 long non-coding RNAs (lncRNAs) using targeted sequencing. Long non-coding RNAs are a type of RNA molecule that do not code for proteins but can play significant roles in regulating gene expression and disease processes.
The study design
Key Findings: Age and LEF1-AS1
The analysis identified two key predictive features for in-hospital mortality: the patient’s age and a specific long non-coding RNA named LEF1-AS1. By using these features, the researchers developed a machine learning model based on a feedforward neural network classifier. This model showed impressive accuracy, with an Area Under the Curve (AUC) of 0.83 and a balanced accuracy of 0.78 in the discovery cohort, which combined data from three European cohorts involving 804 patients.
Validation and Performance
To ensure the model’s reliability, it was tested on an independent cohort of 482 patients from Canada. The model performed consistently well, confirming its potential as a reliable tool for predicting COVID-19 outcomes.
The Role of LEF1-AS1
Further analysis using Cox regression indicated that higher levels of LEF1-AS1 were associated with a reduced risk of mortality. Specifically, the age-adjusted hazard ratio was 0.54, meaning that patients with higher levels of this RNA had a significantly lower risk of dying from COVID-19. This finding was validated using quantitative PCR, a common laboratory technique, confirming that LEF1-AS1 levels can be measured accurately in hospital settings.
Implications for Healthcare
The development of this predictive model is a significant step forward in the management of COVID-19. By identifying patients at higher risk of severe outcomes, healthcare providers can prioritize and tailor their treatment approaches more effectively. This model, which combines age and LEF1-AS1 levels, provides a robust and practical tool that can be used in hospitals to enhance patient care.
This study demonstrates the power of combining clinical data with advanced machine learning techniques to predict COVID-19 outcomes. The identification of LEF1-AS1 as a key predictive biomarker opens new avenues for personalized healthcare in managing the disease. As we continue to fight the pandemic, such innovative approaches will be crucial in improving patient outcomes and easing the burden on healthcare systems worldwide.
Devaux Y, Zhang L, Lumley AI, Karaduzovic-Hadziabdic K, Mooser V, Rousseau S, Shoaib M, Satagopam V, Adilovic M, Srivastava PK, Emanueli C, Martelli F, Greco S, Badimon L, Padro T, Lustrek M, Scholz M, Rosolowski M, Jordan M, Brandenburger T, Benczik B, Agg B, Ferdinandy P, Vehreschild JJ, Lorenz-Depiereux B, Dörr M, Witzke O, Sanchez G, Kul S, Baker AH, Fagherazzi G, Ollert M, Wereski R, Mills NL, Firat H. (2024) Development of a long noncoding RNA-based machine learning model to predict COVID-19 in-hospital mortality. Nat Commun 15(1):4259. [article]